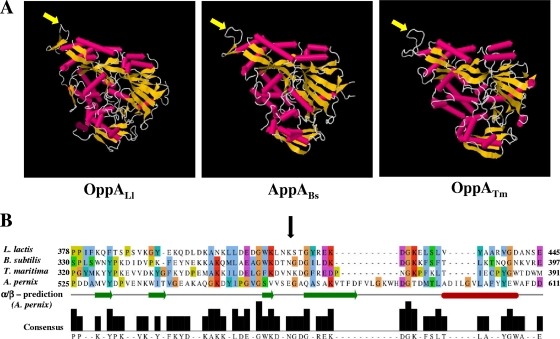
FIG. 2.
(A) Cartoon rocket representation of the three-dimensional (3D) structures of three periplasmic binding proteins: Lactococcus lactis OppA (OppALl) (Protein Data Bank [PDB] accession number 3DRF), AppA from Bacillus subtilis (AppABs) (PDB accession number 1XOC), and Thermotoga maritima OppA (OppATm) (PDB accession number 1VR5). The peptide bond which in OppAAp is subjected to proteolysis is marked by yellow arrows. (B) Multiple-sequence alignment of OppAAp with the binding proteins mentioned above. The alignment was generated by ClustalW and colored according to the ClustalX coloring scheme. The bar consensus graph represents the alignment quality using a BLOSUM62 score based on observed substitutions. The black arrow indicates a common sequence region corresponding to the external loop, which is nicked in OppAAp.