Abstract
The in vivo expression levels of four rRNA promoter pairs (rrnp1p2) of Bacillus subtilis were determined by employing single-copy lacZ fusions integrated at the amyE locus. The rrnO, rrnJ, rrnD, and rrnB promoters displayed unique growth rate regulation and stringent responses. Both lacZ activity and mRNA levels were highest for rrnO under all growth conditions tested, while rrnJ, rrnB, and rrnD showed decreasing levels of activity. During amino acid starvation induced by serine hydroxamate (SHX), only the strong rrnO and rrnJ promoters demonstrated stringent responses. Under the growth conditions used, the rrn promoters showed responses similar to the responses to carbon source limitation induced by α-methyl glucoside (α-MG). The ratio of P2 to P1 transcripts, determined by primer extension analysis, was high for the strong rrnO and rrnJ promoters, while only P2 transcripts were detected for the weak rrnD and rrnB promoters. Cloned P1 or P2 promoter fragments of rrnO or rrnJ were differentially regulated. In wild-type (relA+) and suppressor [relA(S)] strains under the conditions tested, only P2 responded to carbon source limitation by a decrease in RNA synthesis, correlating with an increase in (p)ppGpp levels and a decrease in the GTP concentration. The weak P1 promoter elements remain relaxed in the three genetic backgrounds [relA+, relA, relA(S)] in the presence of α-MG. During amino acid starvation, P2 was stringently regulated in relA+ and relA(S) cells, while only rrnJp1 was also regulated, but to a lesser extent. Both the relA+ and relA(S) strains showed (p)ppGpp accumulation after α-MG treatment but not after SHX treatment. These data reveal the complex nature of B. subtilis rrn promoter regulation in response to stress, and they suggest that the P2 promoters may play a more prominent role in the stringent response.
The major products of all cellular transcription in Escherichia coli and Bacillus subtilis are rRNA and tRNA, which constitute more than 95% of the total RNA (19, 37). There are 7 rrn operons in E. coli and 10 in B. subtilis, each controlled by tandem (P1 and P2) promoters that are tightly regulated in response to changes in nutritional status and other stress conditions (6, 7, 8, 49). A significant number of studies on the regulation of rrn synthesis in E. coli have been published, while only a few reports focusing on the spore-forming Gram-positive bacterium B. subtilis exist.
In E. coli, the core (−10/−35) region in rrnp1 promoters is preceded by an UP element that increases promoter activity 20- to 50-fold because it is strongly bound by the C-terminal domains of the two α subunits of RNA polymerase (6, 7). Binding sites for the transcription factor FIS, centered at positions −71, −102, and −143 upstream of rrnBp1, increase overall activity by an additional 3- to 8-fold (21). The downstream P2 promoters have been reported to be less active during rapid steady-state growth but are more active at lower growth rates (1a, 12, 21, 30, 31). In addition, it has been suggested that when RNA polymerase molecules initiate at the upstream promoter (P1), the enzyme directly interferes with the ability of additional molecules to initiate from the downstream promoter (P2) as suggested by Adhya and Gottesman (1) in their promoter occlusion model. Several studies indicate that most of the regulation of E. coli rrnp1 and rrnp2 activity is attributable to the changes in the concentrations of the initiating nucleoside triphosphate and the highly phosphorylated guanosine nucleotide (p)ppGpp (4, 11, 14, 30). Amino acid starvation and other nutritional stress conditions lead to the “stringent response,” accompanied by increasing levels of (p)ppGpp via a relA gene-dependent mechanism (6). Changes in the levels of newly synthesized rRNA are influenced by the ATP/GTP concentrations (11, 13, 30). The P2 promoters are less affected by the changes in the concentrations of these signaling molecules than are the P1 promoters (31).
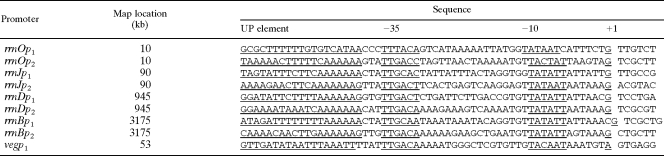
The spore-forming Gram-positive bacterium B. subtilis has 10 rrn operons; 9 are located 1° to 90° on one side of oriC, and 1 (rrnB) is located on the opposite side at 271° on the chromosome (19, 22). The operons are transcribed from the H strand in the same direction as the chromosome replicates (28, 35). Five of the rrn operons (rrnO, rrnA, rrnE, rrnD, and rrnB) have approximately the same organization: tandem promoters P1 and P2-16S gene-spacer-23S gene-5S gene-terminator region. In the closely spaced operons rrnJ-rrnW and rrnI-rrnH-rrnG, there are single promoters (P2) upstream of rrnW and rrnH-rrnG (16, 19, 22). The upstream promoter regions (−36 to −80) are enriched in short A and T tracts, typical for upstream (UP) elements (see Table 2).
Several differences have been noted in the rrn promoter strengths of these operons in both E. coli and B. subtilis (7, 16, 35a). In this paper, we report the differential expression of the tandem promoters P1-P2 of rrnO, rrnJ, rrnD, and rrnB in B. subtilis by employing single-copy lacZ fusions integrated at the amyE locus. We asked whether the operons closest to ori, at map positions 1° (rrnO) and 8° (rrnJ), exhibit higher transcription activity than operons located farther away, at 81° (rrnD) and 271° (rrnB) (2, 19, 35a, 49). We also wanted to determine whether B. subtilis rrnp2 promoters are more active and more highly regulated than rrnp1 promoters, as was shown by others in early studies for rrnO and rrnB (9, 35a, 49). Finally, we inquired whether both rrnp1 and rrnp2 promoters are similarly regulated during amino acid starvation or during carbon source limitation in cell strains differing in their relA backgrounds. Our studies also aimed to evaluate the expression of four rrn operons with respect to their chromosomal positions relative to ori and to establish whether position effects exist in B. subtilis.
MATERIALS AND METHODS
Strains, plasmids, and PCR constructions.
The B. subtilis strains used in this study are listed in Table 1. Strains IS58 (relA+), IS56 (relA), and L3 [relA(S)] are isogenic except for the relA gene (33). L3 has a stringent response to glucose but not to amino acid starvation and is resistant to 3-amino-1,2,4-triazole (AT) (6, 20). In addition, the relA(S) and relA mutant forms cause the cells to require certain amino acids when grown in minimal medium, as reported for E. coli and Salmonella enterica serovar Typhimurium (6, 17, 39, 41, 50). The transcriptional plasmids pDH32 and pDG268 (9,894 and 9,327 bp, respectively), which contain a promoterless spoVG-lacZ fusion gene, a B. subtilis ribosomal binding site from the spoVG gene, two large regions of the amyE gene, and the cat gene determinants like erm from pC194 (3, 21b, 45), were used. Another plasmid, pDG793, also known as pDG1663 (8,396 bp), was examined; it is similar to pDH32 and pDG268 but instead contains two large regions of thrC and the pE12 erm gene (3, 21a). The lacZ gene in all constructs is transcribed in the opposite direction from the amyE or thrC gene (3, 45). Table 1 lists the constructs derived by cloning rrn promoter fragments into the plasmids described above, and Table 2 lists their respective promoter sequences. The intact cloned promoters from rrnO and veg (plasmids pPW4 and pPW810, respectively) have been described previously (10, 36) and were kindly provided by C. Stewart. Plasmid pAWR118 was constructed by inserting a 1.5-kb PstI rrnJ fragment from B. subtilis strain 168 into pDG268 (Table 1) (51, 52). Plasmid pAWR116 contains a 2.2-kb HindIII rrnD fragment from B. subtilis strain SB25 (51, 52) inserted into pDG268. Plasmid pAWR123 contains a 1.8-kb EcoRI rrnB fragment from pGS227 (15) cloned into plasmid pDG268. The fragments derived from rrnO and rrnB extend to the EcoRI site 0.8 kb into the 16S gene; the rrnJ fragment extends to the PstI site at 0.9 kb of the 16S gene; and the rrnD fragment terminates 0.2 kb into the 23S gene. These rrn-lacZ fusions were transformed into the competent IS58, IS56, and L3 strains (Table 1) using standard techniques, and Cmr amyE (or Ermr thrC) cells were selected for these studies.
TABLE 1.
Bacillus subtilis strains and plasmids used in this study
| Strain or plasmid | Relevant genotype and/or promoter fragment (length) | Source |
|---|---|---|
| Strains | ||
| IS58 | trpC2 lys-3 relA+ | I. Smith |
| IS56 | trpC2 lys-3 relA | I. Smith |
| L3 | trpC2 lys-3 relA(S) | R. Rudner |
| Plasmids | ||
| pDH32 | Ampr CmramyEspoVG-lacZ | D. Henner |
| pDG268 | Ampr CmramyEspoVG-lacZ | P. Stragier |
| pPW810 | pDH32-(P1)-veg (0.48 kb) | C. Stewart |
| pPW4 | pDH32-(P1-P2)-rrnO (1.9 kb) | C. Stewart |
| pAWR118 | pDG268-(P1-P2)-rrnJ (1.5 kb) | A.-M. White |
| pAWR116 | pDG268-(P1-P2)-rrnD (2.2 kb) | A.-M. White |
| pAWR123 | pDG268-(P1-P2)-rrnB (1.8 kb) | A.-M. White |
| pLR501 | pDG268-(P2)-rrnO (339 bp) | D. Liu |
| pLR512 | pDG268-(P1)-rrnO (241 bp) | D. Liu |
| pLR105 | pDG268-(P2)-rrnJ (222 bp) | D. Liu |
| pLR210 | pDG268-(P1)-rrnJ (192 bp) | D. Liu |
| pLR203 | pDG268-(P1-P2)-rrnJ (438 bp) | D. Liu |
TABLE 2.
Sequences of B. subtilis rrnO, rrnJ, rrnD, rrnB, and veg P1 and P2 promoters used in this study
The individual promoters of rrnO and rrnJ, as well as rrnJp1p2, without the upstream activation sequences (UAS) were created by PCRs using standard techniques. The primers used, along with their restriction enzyme termini, are listed in Table 3 (27). The rrn promoter fragments generated by PCR were cloned into pDG268 and are listed in Table 1. The rrnOp1 fragment is a 241-bp HindIII-BamHI fragment extending from −186 to +40 with respect to the +1 transcription start site; rrnOp2 is a 339-bp BamHI fragment extending from −84 to +243; rrnJp1 is a 192-bp EcoRI-HindIII fragment extending from −162 to +20; rrnJp2 is a 222-bp HindIII-BamHI fragment extending from −82 to +133 (27); and rrnJp1p2 is an EcoRI-BamHI fragment extending from −162 of P1 to +133 of P2. The DNA sequences of the rrnJ PCR products were confirmed.
TABLE 3.
Primers used for PCRs
| Primer name | Sequencea | Restriction site(s) | Locationb | Promoter fragment(s) generatedc |
|---|---|---|---|---|
| RR15-71 | GGGGGATCCGCTCGACTTGCATGTAT | BamHI | Downstream | rrnJp2, rrnJp1p2, rrnOp2 |
| RR15-72 | GGGAAGCTTGCCGCTAAACAAGGCG | HindIII | Upstream | rrnJp2 |
| RR15-73 | GGGGAAGCTTCCCCTTCTATTCGCGAT | EcoRI | Upstream | rrnJp1, rrnJp1p2 |
| RR15-74 | GGGAAGCTTCGCCTTGTTTAGCGGC | HindIII | Downstream | rrnJp1 |
| RR15-77 | CCGGATCCTGCAGACACAAGCATGACC | BamHI, PstI | Upstream | rrnOp2 |
| RR15-78 | CCGGATCCTAGTCATAATGGTCATGC | BamHI | Downstream | rrnOp1 |
| RR15-79 | GGGAAGCTTCTGCAGGTGCGTCTCAT | HindIII, PstI | Upstream | rrnOp1 |
Regions of primers complementary to target template DNA are underlined; restriction enzyme sites are in boldface.
The priming site of the oligonucleotide with respect to the indicated promoter(s). Upstream primers are sense sequences, and downstream primers are antisense sequences.
Some primers were used to generate several different promoter fragments.
Bacterial growth conditions and in vivo labeling.
Liquid cultures of B. subtilis with or without integrated rrn-lacZ fusions were grown in a fast complex medium composed of 2.5% veal infusion broth and 0.5% yeast extract (VY; Difco) (25). The basal medium (MM) used for growth rate experiments was Spizizen minimal salts (2a) supplemented with 50 μg/ml of l-tryptophan and 100 μg/ml of l-lysine. To achieve different growth rates, the basal medium was supplemented with the following: either 0.5% glucose and 1% sodium glutamate (MM1), 1% sodium succinate, 0.05% yeast extract, and 0.02% vitamin-free Casamino Acids (MM2), or 1% sodium acetate, 0.05% yeast extract, and 0.02% vitamin-free Casamino Acids (MM3). Overnight cultures of the various strains containing integrated rrn-lacZ fusions were centrifuged, washed, and resuspended 1:25 in the same medium or dilution salts, followed by inoculation of the cells into a 250-ml side-arm flask with an additional dilution of 1:10; hence, the cells were diluted 1:250 with the same fresh, prewarmed medium or one of the other types of growth medium and were then shaken at 37°C. During their logarithmic growth, the cells were monitored on a Klett-Summerson spectrophotometer equipped with a red filter. Samples for β-galactosidase assays and for RNA extraction were withdrawn, inactivated with 0.05 M sodium azide, rapidly cooled to 0°C, and then processed. For stringent-response measurements, cultures were grown in MM1 and samples were withdrawn at varying times following the addition of serine hydroxamate (SHX) at 2 mg/ml (47) or α-methyl glucoside (α-MG) at 1% (23). For message decay assays, samples were withdrawn at different times after the addition of rifampin (150 μg/ml) at Klett readings of 100 to 120.
(p)ppGpp measurements.
A low-phosphate Tris-glucose medium described previously (17, 33, 46) was used to label the pool of nucleotides. The medium was supplemented with 100 μg/ml of the l-amino acids lysine, proline, glycine, alanine, glutamic acid, aspartic acid, and arginine and with 40 μg/ml of cysteine, methionine, tyrosine, tryptophan, and phenylalanine to relieve the cells from the low-phosphate condition. Cultures were labeled with [32P]phosphoric acid (50 to 100 μCi/ml) for one generation (1 h), followed by treatment with either SHX, α-MG, or rifampin as described above. Samples (100 μl) were withdrawn at 0, 5, 10, and 20 min and were mixed with 20 μl 13 M formic acid before freezing. The samples were centrifuged for 5 min, and 10 to 20 μl of the supernatants was applied to polyethyleneimine (PEI)-cellulose plates (Brinkmann Instruments) for separation by thin-layer chromatography (TLC) of the phosphorylated guanosine nucleotides in 1.5 M KH2PO4 as described previously (17, 33, 46). Radioactively labeled nucleotides were visualized by autoradiography. The relative concentrations of pppGpp, ppGpp, and GTP were determined by densitometry using a Zeineh soft scanning densitometer (model SL-DNA; Biomed Instruments). Actual nucleotide concentrations were determined by cutting out the appropriate areas of the chromatogram and measuring the radioactivity by liquid scintillation counting. Background radioactivity was corrected for by counting appropriate blank regions of the chromatogram.
Measurements of β-galactosidase activity.
Frozen cell pellets were resuspended in 0.9 ml Z-buffer, solubilized with four drops of toluene, and assayed for β-galactosidase according to published procedures (29). In order to calculate Miller units, Klett readings of the sampled cell suspensions were converted to optical densities at 600 nm.
RNA isolation and quantification.
RNA was isolated using a modified procedure described for E. coli (32). Samples (2.0 to 4.0 ml) were removed from cultures grown in MM1, MM2, or MM3 medium. These samples were diluted with an equal volume of ice-cold TMA buffer (50 mM Tris-HCl [pH 7.4], 1 mM MgCl2, 10 mM NaCN3) and were then centrifuged, and the pellets were immediately resuspended in 2.0 ml of freshly made lysis buffer (2.0 mg/ml lysozyme, 2 U/ml of RNase-free DNase [RQ1; Promega], 10 mM Tris-HCl [pH 8.0], 1 mM EDTA) and were placed on ice for 10 min. This was followed by the addition of 0.5 ml of 1 M NaCl, 50 mM EDTA (pH 8.0), and 2.5% sodium dodecyl sulfate (SDS). Samples were then placed in boiling water for 30 s. After phenol extraction and ethanol precipitation, the RNA was resuspended in sterile water, and the intactness of the preparations was judged by visualization of rRNAs in ethidium bromide-stained agarose gels. The RNA concentration was determined colorimetrically by the orcinol method (43).
Northern slot blot hybridization and primer extension analysis.
To measure lacZ mRNA levels, 5 to 10 μg of total RNA was loaded per slot of a Minifold II slot blotter (Schleicher & Schuell) fitted with Nytran nylon membranes. The membranes were baked, prehybridized, hybridized, and washed as described previously for Northern blot analysis (48). A lacZ probe was generated from pDEB1 (44) by labeling with [α-32P]dCTP using a random primer extension kit as directed by the supplier (USB, Cleveland, OH). Specific activity was consistently 1 × 108 to 3 × 108 cpm/μg of DNA. Membranes were exposed for 17 h to X-Omat AR film (Eastman Kodak Co., Rochester, NY) with intensifying screens at −70°C. Hybridization intensity was determined by scanning autoradiograms using the Zeineth soft scanning densitometer. Primer extension reactions were performed by modification (48) of a procedure previously described for E. coli (12, 32). Briefly, 75 μg of total RNA was mixed with 8 to 10 ng of a 5′-end-labeled primer in 0.1 M KCl-0.05 M Tris-HCl (pH 8.3). This mixture was denatured by heating for 1 min at 90°C, followed by 2 min at 60°C, and was then placed on ice for 15 min to anneal. Then 5XRT buffer (0.25 M Tris-HCl [pH 7.9], 0.2 M KCl, 0.036 M magnesium acetate, 0.01 M dithiothreitol [DTT]), 1 mM deoxynucleoside triphosphates (dNTPs), and 2 U/μl RNasin (Promega) were added to a final concentration of 1×. The primer extension reaction was initiated by the addition of 20 U of avian myeloblastosis virus (AMV) reverse transcriptase (Molecular Genetic Resources); the reaction mixture was incubated for 1 h at 45°C; and the reaction was stopped by the addition of an equal volume of dye mixture (0.1% bromophenol, 0.1% xylene cyanol in deionized formamide). The mixture was heated for 3 min at 100°C, and the primer extension products were separated by electrophoresis on a 6% acrylamide-urea sequencing gel and were visualized by autoradiography. The primer extension products were quantitated by densitometry using the Zeineth soft scanning densitometer. The two primers used were (i) 5′ TGC AGG CCC TAG TTT GAC TGA CTA C 3′, complementary to the unique sequence of rrnO at −256 to −221 (34), and (ii) 5′ TCA GTA ACT TCC ACA GTA GTT CAC CAC CTT 3′, complementary to the spoVG-lacZ junction between the SalI and BamHI sites at pDH32 positions 3771 to 3741 (45). The primers were purified by gel filtration using a 10-ml Sephadex G25X Fine column, and the peak fractions were concentrated by evaporation. The purified primers (100 ng) were then end labeled with 40 μCi of [γ-32P]ATP in hybridization solution (5× kinase buffer, 0.5 M Tris-HCl [pH 9.5], 50 mM MgCl2), 50 mM DTT, and 5 to 10 U of T4 polynucleotide kinase (Boehringer Mannheim). The reaction mixture was incubated at 37°C for 30 min, followed by purification through a Quick Spin G-25 Sephadex column, and was stored at a concentration of 70 to 100 μg/ml.
mRNA half-life determinations.
RNA turnover was determined by the decay of hybridizable lacZ mRNA following inhibition of transcription by rifampin as described above. mRNA half-lives (t1/2) were quantitated by analyzing autoradiograms of Northern slot blots using a densitometer as described above (5, 48).
RESULTS
Do the four intact rrn promoters inserted into the amyE gene display unique levels of expression as a function of growth rate?
To evaluate the intrinsic promoter strength of a representative sample of rRNA gene sets, four promoter-bearing fragments were inserted upstream of the spoVG-lacZ fusion of either pDH32 or pDG268. Fragments from the rrnO, rrnJ, and rrnB operons contained 16S sequences of similar lengths (0.8, 0.9, and 0.8 kb, respectively), while the rrnD fragment included a longer segment of 1.67 kb. The control promoter vegp (0.48 kb), which is constitutively expressed during vegetative growth, was also included (10, 36). The five plasmids bearing the rrn- or veg-lacZ fusions were linearized, and each was transformed into strain IS58 (relA+). The bacteria that harbored integration events of the single-copy intact rrn- or veg-lacZ fusions into the amyE gene were isolated as chloramphenicol-resistant amyE colonies that were blue on minimal agar plates supplemented with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). Colonies that arose by a Campbell-like insertion and yielded the amyE+ phenotype (3, 45) were not chosen for these studies. The cloned rrn-spoVG-lacZ fragments were oriented in the opposite direction from the other functional parts of the plasmids and their drug resistance genes; hence, readthrough of these sequences was unlikely, and only the cloned promoters directed lacZ expression.
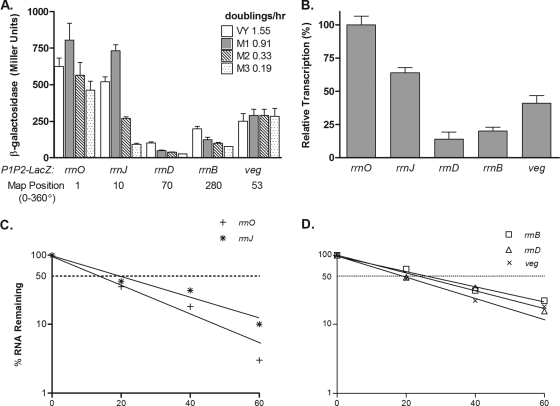
The B. subtilis transformants carrying the various rrn-spoVG-lacZ fusions were grown in four different media to achieve growth rates from 0.2 to 1.6 doublings per h. These cultures were then assayed for β-galactosidase activities. As shown in Fig. 1A, each promoter exhibited a distinct expression level, with rrnO showing the highest expression, as in our earlier studies (51). In VY medium, the relative expression levels of the promoters were 100, 84, 16, 33, and 40% for rrnO, rrnJ, rrnD, rrnB, and veg, respectively. The levels of β-galactosidase expression driven by rrnO were the highest in all media tested, while those for rrnJ were high in complex medium (VY) and in MM1 (glucose) and showed a reduction in promoter strength at lower growth rates. The rrnD and rrnB promoters were weak at all growth rates, while veg displayed a constant activity level under all growth conditions (Fig. 1A). We have repeatedly noticed that cultures growing in VY medium tend to exhibit premature lysis, which occurs as the cells are centrifuged; hence the lower levels of β-galactosidase seen for rrnO and rrnJ in VY than in MM1 medium (Fig. 1A). At the thrC locus at 284° on the map (2), the same rrnJ and rrnD promoter fragments exhibited further decreases from their levels at the amyE locus (data not shown).
FIG. 1.
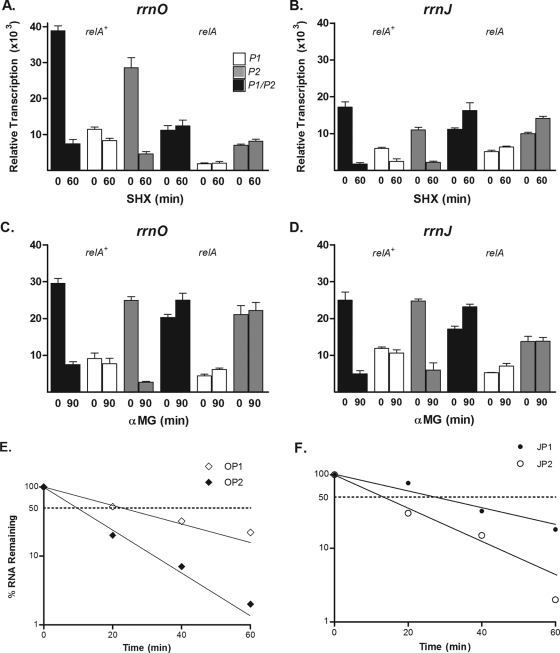
Relative promoter strengths of four rrn operons and the veg gene integrated into the amyE locus. (A) β-Galactosidase activity measured in strains with lacZ-rrn promoter fusions with rrnO, rrnJ, rrnB, rrnD, and veg integrated at the amyE locus cultured in the indicated media to produce different growth rates. One-milliliter samples from the growing cultures were removed at a reading of 100 Klett units and were assayed for β-galactosidase activity by the method of Miller (29) as described in Materials and Methods. (B) The steady-state transcript levels driven by the indicated promoters were determined by Northern slot blotting. Five- and 10-μg portions of total RNA prepared from B. subtilis relA+ strains grown in MM1-glucose were loaded into individual slots and hybridized with an excess of a 32P-labeled lacZ probe. Autoradiograms were analyzed by densitometry, and the values shown are expressed relative to the signal from rrnO. (C and D) The stabilities of rrn-lacZ fusion transcripts initiating at the indicated promoters were determined by Northern slot blot hybridization of a lacZ probe to total RNA prepared at the indicated times (minutes) after rifampin treatment. The half-life was calculated by plotting the relative transcript levels obtained from the densitometric measurements of the autoradiograms versus time on semilog graph paper.
Do the lacZ-mRNA levels correlate with the enzymatic activities of β-galactosidase?
Northern slot blotting was used to determine the transcript levels of lacZ mRNA from cultures grown in MM1, and the results are presented in Fig. 1B. The relative transcript levels (100, 64, 14, 20, and 41% for rrnO, rrnJ, rrnD, rrnB, and veg, respectively) correlated well with the β-galactosidase activity levels (100, 90, 7, 15, and 36%) (Fig. 1A), verifying the heterogeneity in promoter strength among the rrn operons of B. subtilis (42).
Are the half-lives of lacZ mRNAs transcribed from the different rrn promoters similar?
To ensure that changes in lacZ mRNA levels and β-galactosidase activities were not due to differential message stability, the half-lives of the various mRNAs transcribed from intact rrnO, rrnJ, rrnD, rrnB, and veg promoters after standard rifampin treatment were determined by Northern slot blot hybridization using the lacZ probe. As shown in Fig. 1C and D, the half-lives of the reporter mRNAs differed over a small range. lacZ mRNA driven by rrnO, rrnJ, rrnB, rrnD, or veg had a t1/2 of 14.4, 19.1, 26.4, 22.4, or 19.5 min, respectively. Thus, the large differences in mRNA transcript levels and β-galactosidase activities controlled by these rrn promoters must be due to intrinsic promoter activity (42). Our results are consistent with the 2003 report of Hambraeus et al. (18), which showed clearly that more than 30 mRNAs, including both mono- and polycistronic transcripts, were extremely stable, with half-lives of ≥15 min. In addition, total-RNA concentrations, determined colorimetrically by the orcinol method (43) during the rifampin treatments, did not increase; hence, the long t1/2 reported above are reliable and were not due to residual RNA synthesis (data not shown).
Is there a differential response of the individual promoter elements expressed under nutritional stress from the native tandem (P1-P2)?
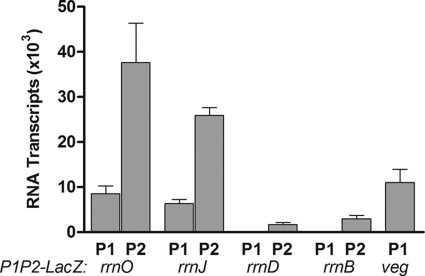
Both of the core rrn promoters (P1 and P2) stimulate the production of the same rRNA; thus, it was of interest to determine their relative contributions to the observed activity of the promoter tandems in integrant strains. These strains were grown in MM1 medium, and promoter activities were compared by quantitative primer extension analysis using a primer that recognizes the spoVG-lacZ fusion transcript. As shown in Fig. 2, transcripts initiating from P2 were more abundant than those starting from P1. In fact, no P1 transcripts from rrnD or rrnB were detectable in this assay. For rrnO and rrnJ, approximately 4-fold fewer transcripts initiated at the P1 promoter than at the P2 promoter. These data suggest that during growth in MM1 with 0.6 doubling per hour, most of the lacZ expression seen in integrant strains is the result of P2 activity. Hence, these data imply that the P2 promoter is dominant over P1 in rrn promoter tandems under these growth conditions.
FIG. 2.
Relative strengths of individual promoter elements as determined by primer extension analysis. Total RNA was isolated from strains containing the indicated promoter-lacZ fusion constructs and was used for primer extension analysis with a 32P-labeled primer that hybridizes to the spoVG-lacZ junction present in the constructs. Primer extension products were separated by gel electrophoresis, and arbitrary values were calculated from densitometric scans of autoradiograms. Transcripts initiating at P1 or P2 were quantitated separately. Each experiment was repeated at least 3 times. Error bars represent the standard errors of the means.
Is there a unique stringent response of solitary PCR-constructed P1 and P2 elements of rrnO and rrnJ during amino acid starvation and carbon limitation?
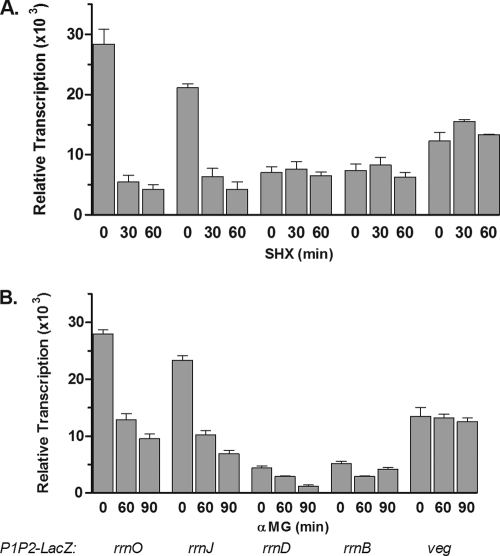
In order to examine stringent regulation of rrn promoter tandems, relA+ strains containing the rrnp1p2-lacZ fusions were stressed with either serine hydroxamate (SHX), a competitive inhibitor of aminoacylation of serine tRNA (47), or α-MG, a competitive inhibitor of glucose uptake (23). The stressed cultures were monitored for their abilities to transcribe lacZ mRNA from the rrn promoter tandems, and the results are shown in Fig. 3 A and B. Within 30 min of SHX challenge, levels of lacZ mRNA transcripts controlled by the strong rrnO and rrnJ promoters decreased dramatically, by 80 to 85%, respectively (Fig. 3A). For SHX (or O-methyl threonine [O-MT]) to act completely effectively, incubation periods of 30, 60, and 90 min have been used for both E. coli and B. subtilis cultures (9, 17, 38, 47). In contrast, the two weak promoters of rrnD and rrnB, as well as the veg promoter, failed to respond to SHX treatment; levels of lacZ mRNA that were not insignificant remained essentially unchanged (Fig. 3A).
FIG. 3.
Effects of nutritional stress on intact rrn promoters. Cultures of B. subtilis containing the indicated promoter-lacZ fusion constructs were grown in MM1-glucose medium and were treated either with serine hydroxamate (A) or with α-methyl glucoside (B) for the indicated times. Total RNA extracted from these cultures was analyzed by Northern slot blotting using a lacZ probe. The resulting filters were subjected to autoradiography and were then scanned. The values shown are average arbitrary densitometric units determined from at least 3 experiments. Error bars represent the standard errors of the means.
Similar measurements with the rrn-lacZ fusions in the same strains after exposure to α-MG are reported in Fig. 3B. As shown, transcription by the strong rrnO and rrnJ promoters decreased, while the rrnB promoter showed a small decline of only 60 to 70% in lacZ mRNA transcription after 90 min of α-MG treatment. As with SHX treatment, the veg promoter failed to respond to carbon source limitation. These data, taken together, demonstrate that whereas all rrn promoters are downregulated during carbon source limitation, only the stronger rrnO and rrnJ promoters can be modulated by amino acid starvation. It is also clear that these two types of stress elicit different cellular responses.
Do the individual rrn promoter elements P1 and P2 respond to nutritional stress?
The roles of the individual rrn promoters within the P1-P2 tandem during the stringent response were investigated using multiple approaches. First, quantitative primer extension analysis was performed on B. subtilis strains differing only in their relA phenotypes [i.e., relA+, relA, relA(S)]. The results, shown in Fig. 4A, suggest that under these conditions, most of the transcripts initiated at the endogenous rrnOp2 promoter, while fewer were from the rrnOp1 promoter, and were stringently regulated in response to both SHX and α-MG treatments in a relA+ background. As expected, neither rrnO promoter element was inhibited by the same treatments in a relA background. Interestingly, in the relA(S) strain, α-MG regulated rrnOp2, but SHX did not. This response is most likely related to the uneven cellular production of highly phosphorylated guanosine nucleotides between the two treatments (see below).
FIG. 4.
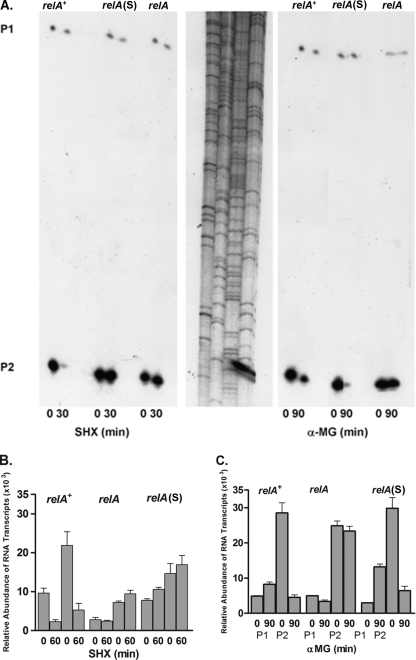
Effects of nutritional stress on individual rrn promoter elements of rrnO. (A) Autoradiogram of a representative primer extension experiment performed using RNA isolated from B. subtilis relA+, relA, and relA(S) strains and an rrnO-specific 32P-labeled primer. Cells were treated with SHX or α-MG for the indicated times prior to RNA extraction. P1 and P2 primer extension products were separated by gel electrophoresis. A sequence ladder of 32P-labeled lambda DNA was included as a size marker for the primer extension products. (B and C) Primer extension analysis using RNA from the different relA strains with the integrated rrnO-lacZ fusion and the spoVG-lacZ primer. Cells were grown in MM1-glucose and were treated with either SHX (B) or α-MG (C) for the indicated times. The relative levels of P1 and P2 transcripts were determined as described for Fig. 2.
The stringent responses of the native P1 and P2 promoter elements of rrnO were then compared to those of the same elements found in the rrnO-lacZ fusion integrated at the amyE locus. Transcript levels were determined by primer extension using the lacZ-specific probe, and the results are shown in Fig. 4B and C. SHX treatment of the relA+-rrnO integrant strain led to similar reductions in rrnO-lacZ transcripts initiating from either rrnOp1 or rrnOp2 (Fig. 4B). SHX had no effect on the levels of these transcripts in either relA- or relA(S)-rrnO integrant strains. In contrast, glucose exhaustion induced by α-MG revealed strong stringent responses of rrnOp2 in the relA+ and relA(S) backgrounds (Fig. 4C). Very low levels of transcripts from the P1 promoters were detected. These did not appear to be regulated by α-MG in any of the integrant strains tested.
Are the stringent responses of solitary rrn promoters also asymmetric when they are expressed from the amyE gene?
Individual P1 or P2 elements from rrnO and rrnJ were amplified by PCR in order to investigate stringent control imposed by the isolated rrn promoters (27). Additionally, an rrnJ fragment containing the promoter tandem and immediate upstream elements (UP), but lacking the upstream activation sequences (UAS), was also isolated. These fragments were cloned into the spoVG-lacZ fusion plasmid, and the resulting constructs were integrated into the amyE locus in both the relA+ and relA backgrounds. Expression of the integrated PCR constructs was compared by RNA dot blots, and the results are shown in Fig. 5A to D. Following the induction of the stringent response by SHX treatment in relA+ integrants, transcript levels controlled by the isolated P2 promoters of rrnO and rrnJ showed dramatic decreases of 84 and 81%, respectively (Fig. 5A and B, shaded bars). The decreases in the levels of lacZ transcripts regulated by the P2 elements were similar to those seen when both the P1 and P2 elements were present (Fig. 5A and B, filled bars). The levels of lacZ transcripts initiating at the isolated P1 promoters of rrnO and rrnJ decreased by only 28 and 59%, respectively (Fig. 5A and B, open bars) As expected, no downregulation of these constructs was observed when they were integrated into a relA strain (Fig. 5A and B).
FIG. 5.
Responses of separated promoter elements to nutritional stress. Wild-type relA+ cells containing the indicated lacZ promoter fusion constructs were grown in MM1 and were treated with SHX (A and B) or with α-MG (C and D) for the indicated times. The relative activities of the promoter fragments of rrnO and rrnJ were measured by densitometric scans of autoradiograms from RNA slot blots before and after treatments. The values were calculated as described in the legend for Fig. 3. (E and F) The chemical half-lives of lacZ mRNAs transcribed from the isolated P1 or P2 elements of rrnO and rrnJ were determined and plotted as described in the legend for Fig. 1C and D.
Glucose exhaustion induced by the addition of α-MG produced a greater differential effect on the expression of lacZ mRNA controlled by the solitary promoters. The P2 promoters responded to glucose starvation in the relA+ strains, demonstrating 89% and 76% inhibition of lacZ transcription from rrnOp2 and rrnJp2, respectively (Fig. 5C and D, shaded bars). As in the case of SHX treatment, the decreases in the levels of lacZ transcripts regulated by the P2 elements were similar to those seen when both the P1 and P2 elements were present (Fig. 5C and D, filled bars). In contrast, the solitary P1 promoters were totally insensitive to carbon deprivation. As expected, no constructs were downregulated in a relA background.
lacZ mRNA levels were measured after rifampin treatment in order to establish whether transcripts initiating at the isolated P1 and P2 promoter elements had different half-lives. The results, shown in Fig. 5E and F, actually indicate that transcripts directed by the strong P2 promoters of rrnO and rrnJ have shorter half-lives than those initiating at P1. Transcripts starting at the isolated rrnOp2 and rrnJp2 had half-lives of 9 and 13 min, respectively. The respective P1 promoters gave rise to transcripts with half-lives of 25 and 27 min. These data suggest that, due to differing half-lives, the increases in the levels of transcripts driven by isolated P2 versus P1 are underestimated, and thus, it is possible that the differential stringent regulation is even more pronounced than that observed. Similarly, the constructed individual promoter elements (P1 versus P2) of rrnO and rrnJ had different decay rates, where the weak P1 promoters had considerably longer t1/2 than the active P2 promoters. In MM1 medium, the lacZ mRNAs transcribed from rrnOp1 and rrnOp2 had t1/2 of 24.7 and 8.8 min, respectively, and the t1/2 of rrnJp1 and rrnJp2 were 27.1 and 12.9 min, respectively (Fig. 5E and F).
Does the stringent control of the P2 promoter correlate with the formation of highly phosphorylated guanosine nucleotides?
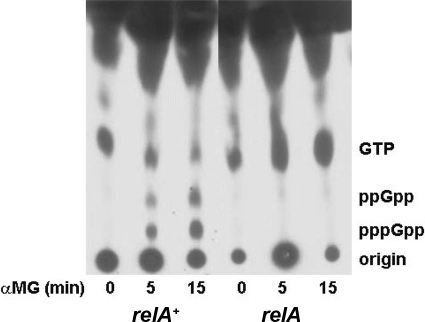
The accumulation of highly phosphorylated guanosine nucleotides correlates well with the induction of the stringent response. Since the rrnp1 and rrnp2 promoters demonstrated differential responses to SHX versus α-MG, it was of interest to assess the status of phosphorylated guanosine nucleotides after exposure to these two agents. As shown in Fig. 6 and Table 4, carbon limitation induced by α-MG resulted in the accumulation of (p)ppGpp in the B. subtilis relA+ strain [results for the relA(S) strain are not shown here but were reported previously (17)] but not in the relA mutant. This considerable increase in ppGpp and pppGpp accumulation was accompanied by a simultaneous decline in the level of GTP to 22 to 25% of the starting levels of GTP found in untreated cultures (Fig. 6 and Table 4). The use of SHX to induce amino acid starvation resulted in larger accumulations of (p)ppGpp both in E. coli and in B. subtilis (6, 17). In the relA+ strain, the levels of pppGpp and ppGpp increased dramatically following 15 min of SHX exposure, while levels of radiolabeled GTP decreased to 20% of the starting amount. In the relA and the relA(S) strains, the levels of GTP did not decrease, and no accumulations of guanosine polyphosphates were detectable (Table 4) (17, 41).
FIG. 6.
Accumulation of (p)ppGpp in B. subtilis relA+, relA, and relA(S) strains during nutritional stress. The indicated strains were grown in low-phosphate medium, followed by a 1-h labeling with [32P]phosphoric acid. Labeled cells were stressed with SHX or α-MG, and samples were removed at the indicated times and were processed for thin-layer chromatography on PEI plates as described in Materials and Methods. The autoradiogram shows results for a representative carbon source starvation experiment.
TABLE 4.
Relative GTP levels before and after treatments and accumulation of (p)ppGpp in three genetic backgroundsa
| Treatment and strain | Treatment time (min) | GTP level (%) | Accumulation (nmol/A690) |
|
|---|---|---|---|---|
| ppGpp | pppGpp | |||
| SHX | ||||
| relA+ | 0 | 100 | 0 | 0 |
| 15 | 20 | 0.58 | 0.75 | |
| relA | 0 | 100 | 0 | 0 |
| 15 | 170 | 0 | 0 | |
| relA(S) | 0 | 100 | 0 | 0 |
| 15 | 83 | 0 | 0 | |
| αMG | ||||
| relA+ | 0 | 100 | 0 | 0 |
| 15 | 22 | 0.42 | 0.43 | |
| relA | 0 | 100 | 0 | 0 |
| 15 | 86 | 0 | 0 | |
| relA(S) | 0 | 100 | 0 | 0 |
| 15 | 25 | 0.11 | 0.16 | |
The levels of GTP were calculated by densitometric scans of autoradiograms, The absolute levels of (p)ppGpp were determined by excision of the appropriate spots from TLC plates, followed by scintillation counting, performed as in a previous study (17).
DISCUSSION
Although there is little variation in the primary sequences of the 10 rRNA genes in B. subtilis or the 7 in E. coli, they respond differently to a variety of physiological conditions. To date, we have examined the activities of four specific rRNA promoters as single-copy integrants into the amyE locus and of two as integrants into the thrC locus. Previously, we constructed 6 of the 10 rrn genes as single-copy insertional plasmids that integrated at their native loci (52). These studies were carried out with fusions of plasmid pDEB1 that originated from pCED6, containing a promoterless E. coli lacZ gene with translational trp sequences of E. coli and a selectable Cmr marker (44). The presence of the E. coli translational sequences in the plasmids used resulted in lower levels of β-galactosidase (20 to 120 Miller units), yet the same gradient of promoter strength (i.e., rrnO, rrnA, rrnW, rrnE, rrnD, and rrnB) was observed (51). The use of another fusion system (rrn-bgaB) from the thermostable organism Bacillus stearothermophilus (20a), integrated at the amyE locus, resulted in a similar gradient of promoter strength (i.e., rrnO, rrnE, and rrnD) (8). In B. subtilis and E. coli, the expression hierarchy of β-galactosidase or chloramphenicol acetyltransferase (CAT) activities from fusions in various media correlates with their original genomic locations relative to oriC (7, 8, 19, 51). Even at the amyE locus, located at 25° on the B. subtilis map (2), the promoters of rrnO and rrnJ, located at 1° and 10°, respectively, clearly retained their high activities, while the promoters of rrnD and rrnB, located at 70° and 280°, respectively, remained weak (2) (Table 2; Fig. 1A and B). Moreover, at their native locations, rrnD and rrnB are linked to the two largest tRNA gene clusters, trnD (16 tRNA genes) and trnB (21 tRNA genes) (19). These clusters are symmetrically situated downstream of these low-expressing operons and are located farthest away from the oriC region (2, 16, 40). Since these tRNA genes are actually part of the rRNA transcriptional unit (16), it has been suggested that this arrangement may offer an explanation for the differences in strength and could define a temporal mechanism of regulation for the 10 ribosomal operons in the endospore-forming bacterium B. subtilis (35a, 40). Our results do not agree with those of another report dealing with similar lacZ rRNA promoter fusions integrated at the amyE locus, which showed that the two elements (P1 and P2) have similar expression levels and that rrnp1 promoters from rrnO or rrnB are essentially as regulated as rrnp2 promoters (see Fig. 1E in reference 24). Krásný and Gourse (24) did not detect the asymmetry in expression between these two promoters reported here and by Okamoto and Vold (35a). The differences in the findings could be related to the lengths of the promoter-bearing fragments. Specifically, our fragments (Table 1) and those used by Okamoto and Vold (35a) were considerably longer (and included additional “UP” sequences) than those used by Krásný and Gourse, which were 58 to 59 bases long (24). In 2006, Koga et al. (23a) reported similar results showing differential transcription of rrn during spore development in B. subtilis, where the P1 and P2 promoter activity levels of rrnO, rrnE, and rrnD were greater in rich medium than in poor medium.
In B. subtilis and E. coli, nutritional limitations trigger the stringent response, which results in the cessation of stable RNA synthesis (6, 19). We have compared the effects of SHX and α-MG on the expression of five promoter fusions in B. subtilis: rrnO-, rrnJ-, rrnD-, rrnB-, and veg-lacZ. Among the promoter fusions tested, only two (rrnO and rrnJ) were stringently regulated, exhibiting 8- to 15-fold decreases in expression upon amino acid or carbon source starvation (Fig. 3A and B and 4A and B). The weak promoters (rrnD and rrnB) and the nonregulated veg promoter did not respond to starvation. In E. coli, all seven operons are stringently regulated, exhibiting 2- to 3-fold decreases following SHX treatment (7). The extent of response in E. coli also varied, with rrnD and rrnE operons having the strongest effect (a 3-fold decrease) and rrnB, rrnC, and rrnH showing a lower decrease of 1.9-fold (7). While transcript stability may differ somewhat by the addition of SHX or α-MG, the relative contributions of the lacZ or CAT fusions in B. subtilis and E. coli should be equal, since they all contain the same portion of the 16S gene. Thus, the differences noted in both bacterial systems are valid measurements of the stringent control of each of the rrn promoters and are not likely to reflect differential effects of mRNA stability (8), as determined by their half-lives (t1/2).
Starvation treatment of B. subtilis relA+ strains led to significant accumulation of (p)ppGpp, while in relA(S) strains, α-MG, but not SHX, caused the appearance of highly phosphorylated guanosine nucleotides (Fig. 6A and Table 4). These findings are contrary to those of Krásný and Gourse (24), who reported no increase in ppGpp accumulation upon α-MG treatment in cells grown in minimal medium containing 0.5 mM KH2PO4, glucose, and 20 amino acids. Our accumulation studies of (p)ppGpp were performed under low-phosphate conditions (0.1 mM KH2PO4) and in the presence of 12 amino acids (see Materials and Methods) (33, 41), and this may account for the different observations.
Despite the relatively significant accumulations of (p)ppGpp induced during α-MG challenge in the relA+ and relA(S) strains (41), transcription from the isolated P1 elements of rrnO and rrnJ persisted, while that from P2 and P1-P2 declined dramatically (Fig. 5C and D). The P1 promoter elements showed small reductions in activity after SHX treatment compared to those of the strong P2 elements (Fig. 5A and B). Since SHX treatment leads to higher levels of (p)ppGpp than does α-MG treatment (Table 4), the behavior of the P1 elements suggests that they can mount a stringent response, but it requires a stronger stimulus, and the P2 element is still the major site of rrn promoter regulation.
The data presented in this study make it clear that B. subtilis rrn operons are heterogeneous with respect to both promoter strength and the stringent response. It also appears that, within the tandem promoters, the P2 element is responsible for most of the regulation seen in the intact promoter. Thus, in contrast to the situation in E. coli, the rrn operons in B. subtilis may have unique, dedicated functions based on the strong differences in stringent responses that we observed.
Acknowledgments
We thank G. Glaser and E. D. Jarvis for critical reading of the manuscript. We also thank undergraduate students Wan Lam, Caroline Barangan, and Sharena Mangal for assisting in some of these studies. We are grateful to Brian Lym, the science reference librarian at Hunter College of CUNY, for his considerable help in providing us with many publications, and Zhong Wang for assisting with computer and network issues. We thank Lisa Weiss for editorial assistance and Nancy Beltrandi for special moral support during the final stages of preparation of this document.
This study was supported by Minority Research Centers in Minority Institutions NIH grant RR0307, by minority Biomedical Research Support Program PHS/NIH grant GM08176-15, and by City University of New York faculty research awards 663131 and 66148.
Footnotes
Published ahead of print on 19 November 2010.
This paper is dedicated to the memory of Inga R. Richter and Steffen R. Buchholz, who were members of our research group and died in an automobile accident (8 June 1988).
REFERENCES
- 1.Adyha, S., and M. Gottesman. 1982. Promoter occlusion: transcription through a promoter may inhibit its activity. Cell 29:939-944. [DOI] [PubMed] [Google Scholar]
- 1a.Afflerbach, H., O. Schroder, and R. Wagner. 1998. Effects of the Escherichia coli DNA-binding protein H-NS on rRNA synthesis in vivo. Mol. Microbiol. 28:641-653. [DOI] [PubMed] [Google Scholar]
- 2.Anagnostopoulos, C., P. J. Piggot, and James A. Hoch. 1993. The genetic map of Bacillus subtilis, p. 425-462. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 2a.Anagnostopoulos, C., and J. Spizizen. 1961. Requirements for transformation in Bacillus subtilis. J. Bacteriol. 81:741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Antoniewski, C., B. Savelli, and P. Stragier. 1990. The spoIIJ gene, which regulates early developmental steps in Bacillus subtilis, belongs to a class of environmentally responsive genes. J. Bacteriol. 172:86-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barker, M. M., and R. L. Gourse. 2001. Regulation of rRNA transcription correlates with nucleoside triphosphate sensing. J. Bacteriol. 183:6315-6323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bechhofer, D. H., and K. H. Zen. 1989. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J. Bacteriol. 171:5803-5811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cashel, M., D. R. Gentry, V. J. Hernandez, and D. Vinella. 1996. The stringent response, p. 1458-1496. In F. C. Neidhardt et al. (ed.), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press: Washington, DC.
- 7.Condon, C., C. Squires, and C. L. Squires. 1995. Control of rRNA transcription in Escherichia coli. Microbiol. Rev. 59:623-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davies, K. M., and P. J. Lewis. 2003. Localization of rRNA synthesis in Bacillus subtilis: characterization of loci involved in transcription focus formation. J. Bacteriol. 185:2346-2353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deneer, H. G., and G. B. Spiegelman. 1987. Bacillus subtilis rRNA promoters are growth rate regulated in Escherichia coli. J. Bacteriol. 169:995-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fukushima, T., S. Ishikawa, H. Yamamoto, N. Ogasawara, and J. Sekiguchi. 2003. Transcriptional, functional and cytochemical analyses of the veg gene in Bacillus subtilis. J. Biochem. 133:475-483. [DOI] [PubMed] [Google Scholar]
- 11.Gaal, T., M. S. Bartlett, W. Ross, C. L. Turnbough, Jr., and R. L. Gourse. 1997. Transcription regulation by initiating NTP concentration: rRNA synthesis in bacteria. Science 278:2092-2097. [DOI] [PubMed] [Google Scholar]
- 12.Gafny, R., S. Cohen, N. Nachaliel, and G. Glaser. 1994. Isolated P2 rRNA promoters of Escherichia coli are strong promoters that are subject to stringent control. J. Mol. Biol. 243:152-156. [DOI] [PubMed] [Google Scholar]
- 13.Gallant, J., G. Margason, and B. Finch. 1972. On the turnover of ppGpp in Escherichia coli. J. Biol. Chem. 247:6055-6058. [PubMed] [Google Scholar]
- 14.Gourse, R. L., T. Gaal, M. S. Bartlett, J. A. Appleman, and W. Ross. 1996. rRNA transcription and growth rate-dependent regulation of ribosome synthesis in Escherichia coli. Annu. Rev. Microbiol. 50:645-677. [DOI] [PubMed] [Google Scholar]
- 15.Green, C. J., G. C. Stewart, M. A. Hollis, B. S. Vold, and K. F. Bott. 1985. Nucleotide sequence of the Bacillus subtilis ribosomal RNA operon rrnB. Gene 37:261-266. [DOI] [PubMed] [Google Scholar]
- 16.Green, C. J., and B. S. Vold. 1993. tRNA, tRNA processing, and aminoacyl-tRNA sythetases, p. 683-698. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. American Society for Microbiology, Washington, DC.
- 17.Gropp, M., E. Eizenman, G. Glaser, W. Samarrai, and R. Rudner. 1994. A relAS suppressor mutant allele of Bacillus subtilis which maps to relA and responds only to carbon limitation. Gene 140:91-96. [DOI] [PubMed] [Google Scholar]
- 18.Hambraeus, G., C. von Wachenfeldt, and L. Hederstedt. 2003. Genome-wide survey of mRNA half-lives in Bacillus subtilis identifies extremely stable mRNAs. Mol. Genet. Genomics 269:706-714. [DOI] [PubMed] [Google Scholar]
- 19.Henkin, T. M. 2002. Ribosomes, protein synthesis factors, and tRNA synthetases, p. 313-322. In A. L. Sonenshein, J. A. Hoch, and R. Losick (ed.), Bacillus subtilis and its closest relatives: from genes to cells. ASM Press, Washington, DC.
- 20.Hilton, J., P. C. Kearney, and B. N. Ames. 1965. Mode of action of the herbicide 8-amino 1,2,4 triazole: inhibition of an enzyme of histidine biosynthesis. Arch. Biochem. Biophys. 112:544-547. [DOI] [PubMed] [Google Scholar]
- 20a.Hirata, H., T. Fukazawa, S. Negro, and H. Okada. 1986. Structure of β-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 166:722-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hirvonen, C. A., et al. 2001. Contributions of UP elements and the transcription factor FIS to expression from the seven rrn P1 promoters in Escherichia coli. J. Bacteriol. 183:6305-6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21a.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pE194, a plasmid that specifies inducible resistance to macrolide, lincosamide, and streptogramin type B antibiotics. J. Bacteriol. 150:804-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21b.Horinouchi, S., and B. Weisblum. 1982. Nucleotide sequence and functional map of pC194, a plasmid that specifies inducible chloramphenicol resistance. J. Bacteriol. 150:815-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarvis, E. D., et al. 1988. Chromosomal organization of rRNA operons in Bacillus subtilis. Genetics 120:625-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kessler, D. P., and H. V. Rickenberg. 1963. The competitive inhibition of alpha-methylglucoside uptake in Escherichia coli. Biochem. Biophys. Res. Commun. 10:482-487. [DOI] [PubMed] [Google Scholar]
- 23a.Koga, K., et al. 2006. Construction of Bacillus subtilis strains carrying the transcriptional bgaB fusion with the promoter region of each rrn operon and their differential transcription during spore development. J. Gen. Appl. Microbiol. 52:119-124. [DOI] [PubMed] [Google Scholar]
- 24.Krásný, L., and R. L. Gourse. 2004. An alternative strategy for bacterial ribosome synthesis: Bacillus subtilis rRNA transcription regulation. EMBO J. 23:4473-4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaFauci, G., R. L. Widom, R. L. Eisner, E. D. Jarvis, and R. Rudner. 1986. Mapping of rRNA genes with integrable plasmids in Bacillus subtilis. J. Bacteriol. 165:204-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin, S., and I. Zabin. 1972. Beta-galactosidase. Rates of synthesis and degradation of incomplete chains. J. Biol. Chem. 247:2205-2211. [PubMed] [Google Scholar]
- 27.Liu, D. X. 1998. Differential regulation of the P1 and P2 promoters of ribosomal RNA operons in Bacillus subtilis. Ph.D. thesis. Hunter College of the City University of New York, New York, NY.
- 28.Margulies, L., V. Remeza, and R. Rudner. 1970. Asymmetric template function of microbial deoxyribonucleic acids: transcription of ribosomal and soluble ribonucleic acids. J. Bacteriol. 103:560-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics, p. 352-355. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 30.Murray, H. D., D. A. Schneider, and R. L. Gourse. 2003. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol. Cell 12:125-134. [DOI] [PubMed] [Google Scholar]
- 31.Murray, H. D., and R. L. Gourse. 2004. Unique roles of the rrn P2 rRNA promoters in Escherichia coli. Mol. Microbiol. 52:1375-1387. [DOI] [PubMed] [Google Scholar]
- 32.Nachaliel, N., J. Melnick, R. Gafny, and G. Glaser. 1989. Ribosome associated protein(s) specifically bind(s) to the upstream activator sequence of the E. coli rrnA P1 promoter. Nucleic Acids Res. 17:9811-9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nishino, T., J. Gallant, P. Shalit, L. Palmer, and T. Wehr. 1979. Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J. Bacteriol. 140:671-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ogasawara, N., S. Moriya, and H. Yoshikawa. 1985. Structure and function of the region of the replication origin of the Bacillus subtilis chromosome. IV. Transcription of the oriC region and expression of DNA gyrase genes and other open reading frames. Nucleic Acids Res. 13:2267-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oishi, M. 1969. The transcribing strands of Bacillus subtilis DNA for ribosomal and transfer RNA. Proc. Nat. Acad. Sci. U. S. A. 54:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35a.Okamoto, K., and B. S. Vold. 1992. Activity of ribosomal and tRNA promoters of Bacillus subtilis during sporulation. Biochimie 74:613-618. [DOI] [PubMed] [Google Scholar]
- 36.Ollington, J. F., and R. Losick. 1981. A cloned gene that is turned on at an intermediate stage of spore formation in Bacillus subtilis. J. Bacteriol. 147:443-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paul, B. J., W. Ross, T. Gaal, and R. L. Gourse. 2004. rRNA transcription in Escherichia coli. Annu. Rev. Genet. 38:749-770. [DOI] [PubMed] [Google Scholar]
- 38.Price, V. L., and J. A. Gallant. 1982. A new relaxed mutant of Bacillus subtilis. J. Bacteriol. 149:635-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rudd, K. E., B. R. Bochner, M. Cashel, and J. R. Roth. 1985. Mutations in the spoT gene of Salmonella typhimurium: effects on his operon expression. J. Bacteriol. 163:534-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudner, R., et al. 1993. Two tRNA gene clusters associated with rRNA operons rrnD and rrnE in Bacillus subtilis. J. Bacteriol. 175:503-519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rudner, R., A. Murray, and N. Huda. 1999. Is there a link between mutation rates and the stringent response in Bacillus subtilis? Ann. N. Y. Acad. Sci. 870:418-422. [DOI] [PubMed] [Google Scholar]
- 42.Samarrai, W. 1996. Differential response of Bacillus subtilis ribosomal RNA operons to nutritional stress. Ph.D. thesis. Hunter College of The City University of New York, New York, NY.
- 43.Schneider, W. C. 1957. Determination of nucleic acids in tissues by pentose analysis. Methods Enzymol. 3:680-684. [Google Scholar]
- 44.Schreier, H. J., and A. L. Sonenshein. 1986. Altered regulation of the glnA gene in glutamine synthetase mutants of Bacillus subtilis. J. Bacteriol. 167:35-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shimotsu, H., and D. J. Henner. 1986. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene 43:85-94. [DOI] [PubMed] [Google Scholar]
- 46.Smith, I., P. Paress, K. Cabane, and E. Dubnau. 1980. Genetics and physiology of the rel system of Bacillus subtilis. Mol. Gen. Genet. 178:271-279. [DOI] [PubMed] [Google Scholar]
- 47.Tosa, T., and L. I. Pizer. 1971. Biochemical bases for the antimetabolite action of l-serine hydroxamate. J. Bacteriol. 106:972-982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weir, J., M. Predich, E. Dubnau, G. Nair, and I. Smith. 1991. Regulation of spo0H, a gene coding for the Bacillus subtilis sigma H factor. J. Bacteriol. 173:521-529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wellington, S. R., and G. B. Spiegelman. 1993. The kinetics of formation of complexes between Escherichia coli RNA polymerase and the rrnB P1 and P2 promoters of Bacillus subtilis. Effects of guanosine tetraphosphate on select steps of transcription initiation. J. Biol. Chem. 268:7205-7214. [PubMed] [Google Scholar]
- 50.Wendrich, T. M., and M. A. Marahiel. 1997. Cloning and characterization of a relA/spoT homologue from Bacillus subtilis. Mol. Microbiol. 26:65-79. [DOI] [PubMed] [Google Scholar]
- 51.Widom, R. L. 1988. Ph.D. thesis. Hunter College of the City University of New York, New York, NY.
- 52.Widom, R. L., E. D. Jarvis, G. LaFauci, and R. Rudner. 1988. Instability of rRNA operons in Bacillus subtilis. J. Bacteriol. 170:605-610. [DOI] [PMC free article] [PubMed] [Google Scholar]