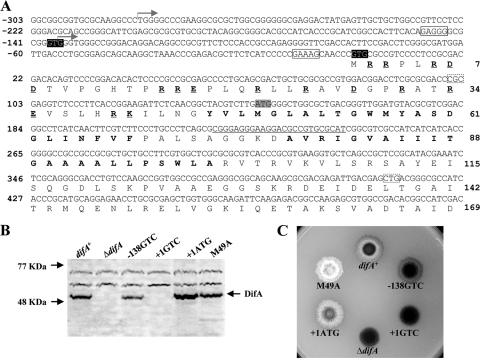
FIG. 1.
Identification of DifA start codon. (A) The upstream and 5′ coding sequence of difA with deduced amino acids. The oligonucleotide used for primer extension (see Fig. S1 in the supplemental material) is complementary to the underlined DNA sequence centered around the codon for D77. The two transcription initiation sites identified by primer extension are marked by two forward arrows, respectively. The two potential GTG start codons for DifA at −138 and +1 are shaded in black, with their consensus ribosomal binding sites boxed with solid lines. The ATG proposed by bioinformatics as the start codon for DifA is shaded in gray. The amino acids in bold are the two predicted transmembrane helices TM1 and TM2. The charged residues preceding TM1 are in bold and underlined. Codons 34 (CGC) and 138 (CTG), boxed by dashed lines, were changed to CGG and CTC as silent mutations to introduce two restriction sites in pWB230 (Table 2). (B and C) Immunoblot analysis (B) and EPS production (C) of strains with mutations of the potential start codons. For immunoblotting, whole-cell lysates from 5 × 108 cells for each strain were separated by SDS-PAGE and probed with anti-DifA antibodies. For EPS analysis, 5 μl of cells at approximately 5 × 109 cells/ml was spotted onto CTT plates containing calcofluor white at 50 μg per ml; the plates were photographed under UV illumination after 5 days of incubation at 32°C. The diameter of the plates shown is ∼9 cm. Strains: difA+, YZ619; ΔdifA, YZ601; −138GTC, YZ709; +1GTC, YZ710; +1ATG, YZ711; and M49A, YZ606.