Figure 1.
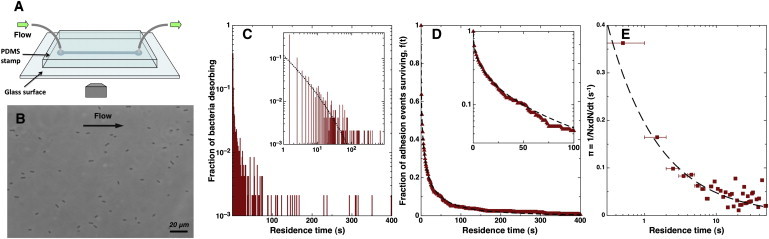
Principle of the experimental protocol and image analysis. (A) The straight channel is imprinted in a PDMS stamp, which is then sealed to a glass slide. Tubing is connected to the device and the flow rate is controlled by means of a syringe pump. (B) Typical field of view with a 40× phase contrast objective. For each measurement focused on the glass surface, 1 image/s was recorded for 25 min. (C–E) Analysis of the residence time distribution at a given flow rate: (C) Distribution of residence times for the population of bacteria. We note that with respect to the quantitative analysis in the Materials and Methods, the vertical axis in panel C is equivalent to P(t) = Ndes(t)/Ntot, where Ndes is the detachment rate of bacteria. (Inset) Log-log plot. (D) Fraction of adhesion events surviving longer than t (or renormalized number of cells on the surface f(t) = N(t)/Ntot with times renumbered so that all cells stick at t = 0). (Dashed line) Best fit with a stretched exponential function (Eq. 4). (Inset) Log-linear plot, which would be linear if the number of bacteria on the surface was decreasing exponentially. (E) Calculated detachment probability π as function of residence time. The data presented on the three graphs were obtained with PA14 wild-type flushed at 6 μL/min in a 400 × 35-μm cross-section channel (wall shear stress = 1.2 Pa). The total number of bacteria counted in this experiment was Ntot = 477. The best fit of f(t) with Eq. 4 corresponds to τ = 5.95 ± 0.11 s and α = 0.39 ± 0.003.These parameters were used to generate the curves superimposed to π(t) and P(t) using Eqs. 9 and 11, respectively.
