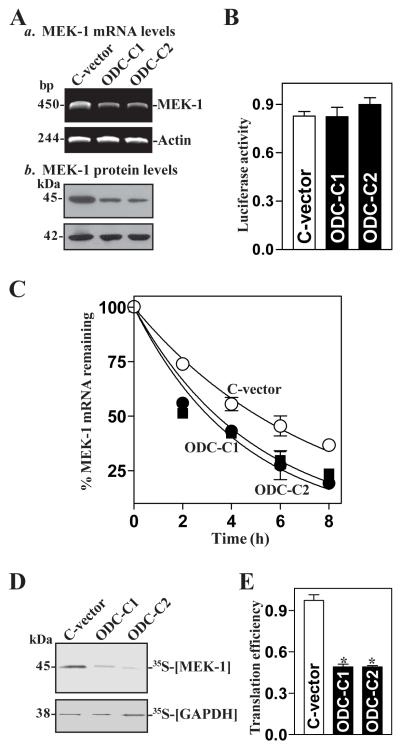
Fig. 2.
Increasing cellular polyamines represses MEK-1 expression. (A) Changes in MEK-1 mRNA and protein in clonal (C) populations of ODC-IEC cells (ODC) and control cells (C-vector). IEC-6 cells were infected with either the retroviral vector containing the sequence encoding mouse ODC cDNA or control retroviral vector lacking ODC cDNA. Clones resistant to the selection medium containing 0.6 mg/ml G418 were isolated and screened for ODC expression. The levels of MEK-1 mRNA and protein were assessed by RT-PCR analysis and Western immunoblotting, respectively. (B) Levels of MEK-1-promoter activity in cells described in A. Luciferase activity was examined 48 h after transfection with pMEK1-luc or the control vector (pGL3). Data were normalized by Renilla-driven luciferase activity and expressed as means ± SE of data from 3 separate experiments. (C) Half-life of MEK-1 mRNA in cells described in A. After cells were incubated with actinomycin D for the indicated times, total cellular RNA was isolated, and the levels of remaining MEK-1 and GAPDH mRNAs were measured by Q-PCR analysis. Values are the means ± SE from triplicate samples. (D) Newly synthesized MEK-1 protein in cells described in A. After cells were incubated with L-[35S]methionine and L-[35S]cysteine for 20 min, cell lysates were prepared and immunoprecipitated by using anti-MEK-1 antibody, resolved by SDS/PAGE, and transferred for visualization of signals by using a PhosphorImager. The translation of housekeeping control GAPDH was measured similarly. (E) Changes in MEK-1 translation efficiency as measured by using pGL3-Luc-MEK1ARE reporter assays in cells described in A. The pGL3-Luc-MEK1ARE or pGL3-Luc (negative control) was cotransfected with a Renilla luciferase reporter, and firefly and Renilla luciferase activities were assayed 24 h thereafter. Luciferase values were normalized to the mRNA levels to obtain translation efficiencies and expressed as means ± SE of data from 3 separate experiments. * p < 0.05 compared with controls.