Abstract
The Biomolecular Interaction Network Database (BIND) is a major source of curated biomolecular interactions, which has been unmaintained for the last few years, a trend which will eventually result in the loss of a significant amount of unique biomolecular interaction information, mostly as database identifiers become out of date. To help reverse this trend, we converted BIND to a standard format, Proteomics Standard Initiative-Molecular Interaction 2.5, starting from the last curated data release (from 2005) available in a custom XML format and made the core components (interactions and complexes) plus additional valuable curated information available for download (http://download.baderlab.org/BINDTranslation/). Major work during the conversion process was required to update out of date molecule identifiers resulting in a more comprehensive conversion of BIND, by measures including number of species and interactor types covered, than what is currently accessible elsewhere. This work also highlights issues of data modeling, controlled vocabulary adoption and data cleaning that can serve as a general case study on the future compatibility of interaction databases.
Database URL: http://download.baderlab.org/BINDTranslation/
Introduction
The Biomolecular Interaction Network Database (BIND) (1–3), is one of the major, freely available molecular interaction (MI) resources, populated over more than 5 years (from 2000 to December 2005 with a few additions in 2006) (4) through detailed manual curation of both high- and low-throughput interactions and automated import of high-throughput interactions. BIND curators, initially a handful, later peaking at more than 40, mined more than 16 000 scientific publications, documented over 200 000 binary interactions and over 3700 biological complexes, from more than 1500 species. From the 16 643 publications curated, 16 438 can be considered low-throughput studies by the BIND team [containing 40 interactions or less (1)], accounting for a third of the total number of interactions (67 789 low-throughput interactions from 206 859 total).
Current access to BIND data is through the BOND web portal (http://bond.unleashedinformatics.com/), run by Thomson Reuters’ Life Sciences Division. While BIND curation ended in 2005, BIND still remains a highly cited publicly available interaction database receiving 117 citations in 2009 alone, comparable to the actively curated and maintained BioGrid (5), HPRD (6) and IntAct (7) interaction databases with 171, 132 and 127 citations, respectively. Although still popular, the gene and protein identifiers contained in BIND are slowly degrading as resources they point to retire or change old identifiers. Further, the original data are not currently available in a generally recognized standard MI format from the official BIND website, though it is available in a simple tab-delimited format, a custom XML format and the Proteomics Standard Initiative-MI (PSI-MI) 2.0 format, an intermediate PSI-MI format that was never officially recognized, both via the website and a download area (http://bond.unleashedinformatics.com/downloads/data/BIND/data/). These factors make it difficult to use the complete BIND database with current software and increase the cost of accessing the knowledge about interactions it contains.
While BIND data in a standard format is not available from the official source, some interaction metadatabases contain and redistribute subsets of BIND data. Databases that incorporate a specific subset of BIND data include Human Annotated and Predicted Protein Interaction (HAPPI) (8), Human Protein Interaction Database (HPID) (9) and UniHI (10) for human interactions, pSTIING (11) for inflammation and cancer, and InnateDB (12) for innate immunity-specific interactions. Others, such as Interaction Reference Index (iRefIndex) (13), Agile Protein Interaction DataAnalyzer (APID) (14) and Michigan Molecular Interactions (MiMI) (15) aim to redistribute and make available a non-redundant set of protein interactions for all species for convenient access over the web or via software tools, like Cytoscape (16). STRING (17), Human Protein–Protein Interaction Prediction (PIPS) (18) and Interologous Interaction Database (I2D) (19) collect and predict interactions and include BIND as an interaction source. In all of these databases, BIND has been a useful source of curated protein interactions as it provides unique interactions that do not overlap with other interaction resources. According to the statistics from iRefIndex (13), there are 25 481 unique BIND interactions (http://wodaklab.org/iRefWeb/statistics/index) among 10 interaction databases.
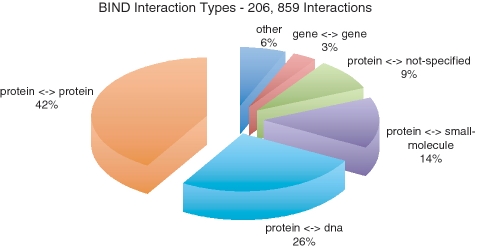
Although, the majority of BIND interactions occur between proteins, as captured in the above databases, BIND also contains many interactions involving RNA, DNA, genes, complexes and small molecules (Figure 1 and Table 1). A large subset of BIND consists of a set of protein—small molecule interactions that were computationally extracted from 3D protein structures from the Molecular Modeling Database (MMDB), originally from the Protein Data Bank (PDB) (20). While BIND was active, the curation team aimed to collect many specific details for interactions and their participants (Table 2), sometimes from additional publications not directly associated with the interaction. Data structures within BIND that contain this specific information include: BIND-place storing the cellular location of the interactions or interactors, BIND-condition detailing experimental conditions, BIND-action listing the chemical actions that can occur in the interaction, BIND-loc storing detailed information about binding sites and BIND-state listing chemical states of the interactors (Table 2). Records also contained detailed comments extracted from publications and their associated figures relating to experimental methods and conditions or original curator written text. Although not standardized to the same degree as translating the information to a controlled vocabulary (CV), curator’s comments are associated to individual BIND data types and are therefore specific to it and offer valuable information pertaining to the interaction. Curated comments generally conform to a defined format in the BIND curation manual (http://bond.unleashedinformatics.com/downloads/data/BIND/docs/curation/BIND_Curation_Training_Manual.pdf). For example, BIND-condition text description should contain ‘An interaction between (species) Molecule A and (species) Molecule B was demonstrated by (experiment)’. Thus, it is useful to make available a comprehensive version of BIND incorporating as much of the curated information as possible in a standard format.
Figure 1.
Interaction types present in BIND.
Table 1.
Main BIND data types and usage statistics
| Element name | Types | Number of instances | |
|---|---|---|---|
| BIND-interaction | 206 859 | ||
| BIND-Interaction_iid | 206 859 | ||
| BIND-Interaction_a | 206 859 | ||
| Complex | 2932 | ||
| DNA | 3020 | ||
| Gene | 10 872 | ||
| Not-specified | 15 635 | ||
| Protein | 143 800 | ||
| RNA | 758 | ||
| Small molecule | 29 842 | ||
| BIND-place | 4955 | ||
| BIND-Interaction_b | 206 859 | ||
| Complex | 645 | ||
| DNA | 54 027 | ||
| Gene | 7337 | ||
| Not-specified | 4005 | ||
| Photon | 291 | ||
| Protein | 131 153 | ||
| RNA | 3706 | ||
| Small molecule | 5695 | ||
| BIND-place | 10 508 | ||
| BIND-Interaction_descr | 206 859 | ||
| BIND-condition | 299 801 | ||
| BIND-place | 739 | ||
| BIND-action | 3272 | ||
| BIND-state (interactor A) | 2027 | ||
| BIND-state (interactor B) | 841 | ||
| BIND-loc | 121 064 | ||
| BIND-descr_intramolecular | 57 | ||
| BIND-Interaction_source | 206 859 | ||
| BIND-pub-set_disputed | 37 | ||
| PubMedId | 254 191 | ||
| BIND-Interaction_division | 206 859 | ||
| BIND-molecular-complex | |||
| BIND-Molecular-Complex_mcid | 3703 | ||
| BIND-Molecular-Complex_descr | 3703 | ||
| BIND-Molecular-Complex_sub-units | 3703 | ||
| Complex | 527 | ||
| DNA | 64 | ||
| Not-specified | 10 | ||
| Protein | 20 764 | ||
| RNA | 78 | ||
| Small molecule | 159 | ||
| BIND-place | 8417 | ||
| BIND-Molecular-Complex_interaction-list | 3703 | ||
| BIND-Molecular-Complex_ordered | 48 = True, 3655 = False | ||
| BIND-Molecular-Complex_source | 3703 | ||
| BIND-pub-set_disputed | 0 | ||
| PubMedId | 4244 | ||
| BIND-Molecular-Complex_division | 3699 |
Table 2.
Fields present in BIND-descr representing mostly unique information with usage statistics
| BIND-descr | Number of instances | Notes | Reason |
|---|---|---|---|
| BIND-condition | 299 801 | ||
| BIND-condition_action | 1740 | Not translated | No good mapping to PSI-MI |
| BIND-condition_bait-condition | 299 801 | Translated | |
| BIND-condition_descr | 294 187 | Translated | |
| BIND-condition_exp-form-a | 163 673 | Translated | |
| BIND-condition_exp-form-b | 250 924 | Translated | |
| BIND-condition_general | 299 801 | List of possible general experimental conditions: in vivo (0), in vitro (1), in situ (2), in silico (3), other (4)—not translated | No good mapping to PSI-MI |
| BIND-condition_genetic-exp | 14 554 | Not translated | Will translate in a future version |
| BIND-condition_negative-result | 26 | Not translated | Will translate in a future version |
| BIND-condition_other-db | 14 | Not translated | Will translate in a future version |
| BIND-condition_site | 63 117 | Not translated | Will translate in a future version |
| BIND-condition_source (individual Pubs) | 292 306 | Translated | |
| BIND-condition_system | 299 801 | Translated | |
| BIND-cons-seq-set | 14 | Not translated | No good mapping to PSI-MI |
| BIND-place | 739 | Translated | |
| BIND-action | 3272 | Not translated | No good mapping to PSI-MI |
| BIND-state (interactor A) | 2027 | Not translated | Partial mapping to PSI-MI |
| BIND-state (interactor B) | 841 | Not translated | Partial mapping to PSI-MI |
| BIND-loc | 121 064 | Not translated | Will translate in a future version |
| BIND-descr_intramolecular | 57 | Translated |
Fields translated in current translation are indicated.
We converted a large fraction of BIND to PSI-MI (version 2.5), the widely accepted and interchangeable standard for representing biomolecular interactions and complexes. PSI-MI is an XML-based schema developed and maintained by the PSI, under the auspices of the Human Proteome Organization (HUPO), for standard representation of MIs (21). This article describes how the core information for every interaction and complex, for every species, in BIND (supplied in BIND XML format) was translated to PSI-MI 2.5 (21) and made available for download. To test the utility of the translation, we validated the results with the PSI-MI validator (22), verified that our translation could be imported to an instance of the PSIQUIC web service for querying interactions and loaded some of the data via this web service into the Cytoscape network visualization and analysis software (16).
Materials and Methods
Data
The BIND data schema describes MIs, complexes and pathways in a high level of detail (23). It is available at http://bond.unleashedinformatics.com/downloads/data/BIND/spec/ and can be browsed online using the ASN.1 browser at http://software.dumontierlab.com/asn-browser/. The BIND schema uses complex data types defined and used by NCBI, such as NCBI-sequence for nucleotide and protein sequences, MMDB which describes 3D molecular structures, and NCBI-pub, which describes publications. BIND curators mainly populated interaction and complex records, and pathways were never populated beyond the initial examples created to demonstrate functionality. Thus, we focused our conversion on interactions and complexes and have chosen PSI-MI 2.5 as the standard format to convert to, as this format covers these data types.
The BIND data was downloaded from http://bond.unleashedinformatics.com/downloads/data/BIND/data/datasets/taxon/xml. The starting data repository used was a 2005 ‘database dump’ of BIND in BIND-XML format, comprising 1589 files (representing 1587 species), with a total size of over 15 GB. Some of these files [for taxids 10 090 (mouse), 4932 (yeast), 562 (Escherichia coli), 7227 (fruit fly) and 9606 (human)] were further split into smaller files for ease of handling during conversion. The files contained 206 859 unique interactions and 3703 complexes. The majority of the interactions are of type protein–protein, protein–DNA, protein-small molecule, protein-unspecified and gene–gene (Figure 1).
BIND is split into divisions for metazoa (all interactions from taxid: 33 208 or its child nodes), fungi (all interactions from taxid: 4751 or its child nodes), taxroot (all interactions not in taxid 33 208 or 4751 or their child nodes), refBIND (reference quality interactions), 3DBP (interaction automatically extracted from structure data containing proteins, RNA or DNA) and 3DSM (interactions automatically extracted from structure data where one of the molecules is a small molecule). All of the interactions found in each of these divisions are also found in the species-specific files, but if an interaction occurs between two molecules where one interactor originates in species X and the other interactor originates in species Y, the interaction will be found in both species files. Although the BIND division files were created such that no file is larger than 2 GB, we used the redundant representation divided according to species, which has many more smaller files, for ease of handling. Therefore, the conversion includes all interactions in all divisions of BIND, but is packaged in a species-specific format that includes duplicate interactions when they exist between species.
BIND was originally represented and stored in the Abstract Syntax Notation One (ASN.1) format. ASN.1 is an International Standards Organization (ISO) data representation format used for data storage by the U.S. National Center for Biotechnology Information (NCBI). BIND modeled MIs and complexes by introducing its own ASN.1 modules that in turn used 20 NCBI ASN.1 modules for representing information about molecules and publications (23). BIND was later converted to XML using NCBI’s ‘datatool’, which can convert any ASN.1 document to XML without loss of information. We used XML as opposed to the original ASN.1 representation as it contained all the information in BIND but is easier to parse using standard XML processing tools.
BIND schema survey and validation
The BIND schema is very complex, including 648 populated unique fields out of over 1600 defined (with 3543 unique paths to those 648 fields). We performed a survey of the use of these fields in the BIND schema before mapping to PSI-MI 2.5 to help prioritize the mapping and conversion. Although the BIND data schema is rich in detail, some fields were never used by curators. For example, for each Bind-gen-place there is an associated start location, end location and description. Of the nearly 20 000 records containing location information, only 79 have both a start and end location defined. Further, the use of some parts of the schema was not standardized, leading to multiple ways to describe the same data. For example, there are four different fields used to store a PubMed ID (PMID) (Pub_medline, Pub_muid, Pub_pmid, PubMedId). Based on this analysis, we chose a subset of frequently used or easy to map fields to convert to PSI-MI format.
Basic interaction conversion
Interaction and complex data were mapped from BIND-XML to PSI-MI version 2.5. Starting from the top level of the BIND schema, a ‘BIND-interaction’ record was converted to a PSI-MI interaction (Supplementary Figure S1). According to the minimal information required to report a protein interaction (MIMIx) (24), each interaction requires a list of interactors and their database identifiers, a basic description of the experiment used to detect the interaction and an associated publication which reports the interaction. We also mapped other data, including curator comments, description text and BIND CV terms, which were translated to the corresponding PSI-MI CV term when possible, as described below. The BIND schema includes a number of custom CVs needed where no standard CVs were available at the time.
Interactor
Each BIND-interaction contains a BIND-object to describe molecules A and B. Each BIND-object was converted to a PSI-MI interactor (Supplementary Figure S2). There is a diverse set of interactions involving combinations of BIND interactor types (protein, RNA, DNA, gene, small molecule, complex, photon, and unidentified interactors), all of which were translated (Figure 1 and Table 1). Two CV types needed to be translated in this process. First, the BIND interactor type was converted to its corresponding PSI-MI interactor type. For example BIND-object_type_id_protein was translated into ‘protein’ (MI:0326 - http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0326&termName=protein) (all CV conversions can be found in Supplementary Table S1). Second, identifier types for BIND interactors were mapped to PSI-MI identifier CV terms, such as Geninfo-id to ‘genbank_protein_gi’ (MI:0851—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0851&termName=protein genbank identifier) and Domain-id to ‘entrezgene/locuslink’ (MI:0477—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0477&termName=entrezgene/locuslink). Last, data from the BIND-object_short_label, BIND-object_descr, and Org-ref was directly transferred to PSI-MI shortLabel, fullName and organism.
The BIND-descr object contains a wealth of information, including experimental conditions (BIND-cond), sequence conservation information (BIND-cons), binding sites (BIND-binding sites), binding actions (BIND-action), binding states (Bind-state-descr) and cellular compartments (BIND-place). We focused on mapping information that was present in PSI-MI (some information is not covered by PSI-MI) and data found in frequently used fields (Table 2), as described below.
Experimental description
We mapped experimental conditions (BIND-cond), as it is the most populated field in the BIND-descr object (Table 2) and is required by MIMIx. Each BIND-cond object was translated to a PSI-MI Experiment Descr object (Supplementary Figure S3). MIMIx states that each experiment should consist of a host system, an interaction detection method and a participant detection method. In a BIND-condition, there is no field specifying the host system of the interaction. Although each interactor is associated with a species, there is no way to infer from this information which species the interaction was detected or modeled in. For this reason, all BIND PSI-MI interactions have no host system defined. BIND uses a custom CV to describe the interaction detection method in BIND-experimental system (Supplementary Table S1). We mapped these to their corresponding PSI-MI vocabulary equivalents. Of the 41 experimental systems described in BIND, 3 are classified by PSI-MI as participant detection methods. Where one of these was included in the BIND record, it was included as the participant detection method. Otherwise, in order to conform to the MIMIx standard and since the majority of records in BIND were curated, we chose to populate the participant detection method as ‘inferred by curator’ (MI:0364—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0364&termName=inferred by curator).
Publication
Each PSI-MI record requires a publication source reference that reports the interaction (bibRef) associated with the experimental description. There are more than 70 places within the BIND schema that can store a publication but many are specific to curated information, like cellular localization, and not the interaction itself. Thus, we only mapped publications from the BIND publication source for the interaction (BIND-interaction_source) and the source for the BIND-condition (BIND-condition_source) with 292 821 and 434 236 total publications listings, respectively (total of 16 643 distinct publications). Each PSI-MI experimental description (which directly corresponds to an individual BIND-condition) contains a bibRef with all the unique publications extracted from the BIND-interaction_source and any additional publications contained in the mapped BIND-condition.
There were 15 782 BIND interaction records without PMIDs but instead had a general citation with the publication title, corresponding to 4732 unique citations. Manual inspection of these determined that this was often due to the record being created prior to the publication of its associated reference, its inclusion in PubMed or, in the case of MMDB derived interactions, a missing publication. In order to update the missing PMIDs, we queried PubMed using batch Entrez with each publication title extracted from the general citation. If PubMed returned an entry matching the query title, the associated PMID was retrieved and updated in the PSI-MI interaction record. Of the 4732 unique citations, we were able to find PMIDs for 1641, which allowed us to update PMIDs in 6080 unique interaction records. A PSI-MI bibRef can contain either an external reference (xref) or an attribute list. For those interactions lacking a PMID, we were unable to find a PMID from the title, the bibRef was populated with an attributelist with the attributes ‘BIND Record’ containing the BIND id and ‘Publication title’ containing the title.
Experimental form
Beyond the requirements of MIMIx, there were additional fields in the BIND-cond that merited translation. The BIND-cond contains descriptions of the experimental forms of both molecules A and B (as BIND-objects). These BIND-objects were translated as described above for interactors but were mapped to PSI-MI participant→experimentalInteractor. These generally contained description of tags added to the molecules, truncated proteins or molecules that originated from a species different than the molecules described to be interacting (e.g. often done in experimental biology when modeling a human interaction in a model organism).
Experimental role
BIND-cond defines the ‘bait-condition’, describing if molecules A or B was used as a bait in the experiment. The bait-condition can be either ‘a-is-bait’, ‘b-is-bait’ or ‘not-applicable’. This information was transferred to participant→experimentalRoleList→experimentalRole as either bait (MI:0496—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0496&termName=bait), prey (MI:0498—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0498&termName=prey) or unspecified (MI:0499—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0499&termName=unspecified%20role) as appropriate.
Cellular location
Additionally, we converted ‘BIND-place’, which represents a cellular location. A BIND-place can be associated with the interactor or the interaction. If the BIND-place was associated with the interactor (‘BIND-object’), it was mapped to PSI-MI interactor→organism→compartment. If the BIND-place was associated with the interaction (‘BIND-descr’), it was mapped to each of the interactors in the interaction as there is no field in the PSI-MI specification describing the interaction location. This mapping created problems when both the interactor and interaction had a place defined as only one compartment can be specified per interactor (12 214 occurrences). In this case, the interactor was annotated with the place specified in the interactor and the additional places were logged but not mapped to the new record.
A BIND-place is composed of four parts, the general place, specific place, source and description. General place, the required element for every BIND-place, consists of a list of 231 places taken from an internally defined CV. Each of the CV terms were mapped to corresponding PSI-MI CV terms (Supplementary Table S1). The specific place contains a Gene Ontology (GO) cellular component term and was mapped directly. The source (e.g. publication) associated with the BIND-place could be the same as the publication describing the interaction, or could be unique when the curator performed additional work to uncover the localizations of the interaction participants. There were only nine instances where the publication associated with the BIND-place was different than the one describing the interaction. Since there is no place in PSI-MI to store this information, it was logged but was not mapped to the new record.
Complexes
There are multiple ways to represent complexes in PSI-MI, including as an interaction containing a set of participants which are part of the complex (flat representation), a set of interactions (participant→interactionRef) that represent the topology of interactions within a complex, or an interaction containing a set of participants with the topology stored as a list of inferredInteractions. BIND only represents complexes as a list of interactions and uses this representation to store both detailed knowledge of complexes including topology, for example, from an X-ray crystal structure, and less detailed knowledge of complexes where topology is not known, but may be inferred, for example, from a proteomics experiment where bait is connected to prey using the spoke model (25). We mapped all BIND complexes to PSI-MI as a flat set of participants with topology stored as a list of inferred interactions. This allows easy access to all BIND complex data. Users wishing to use topological information can access it through the inferred interaction list, though will need to differentiate between detailed and spoke representations.
Additional data
The remaining data not translated from BIND includes sequence conservation information (BIND-cons), binding sites (BIND-binding-sites), binding actions (BIND-action) and binding states (BIND-state). Sequence conservation was not translated due to the limited number of interactions it was present in (total 14 interactions) and because PSI-MI does not model this data. Binding actions and binding states were not translated to PSI-MI as they represent molecular events (e.g. enzymatic reactions) which cannot be stored in PSI-MI. Finally, binding sites were not mapped because most sites were specific to a particular location in the specific sequence reference used in the record. By updating the sequence IDs (described below), the binding site locations in BIND may become invalid if the corresponding sequence has changed, requiring a careful sequence position remapping procedure, which we have not yet implemented.
BIND fields that have no direct mapping to PSI-MI were mapped to generic attributes in the PSI-MI attributeList, either at the interaction or participant levels. For instance, we included attributes ‘BIND interaction division’ to contain the BIND division, ‘BIND curator compartment description’ containing curator comments associated with the BIND-place, ‘BIND record’ containing the original BIND identifier, ‘Publication title’ for publications that did not have PMIDs and ‘Complex Number of Subunits’ containing the number of subunits in a complex.
The PSI-MI ID attribute for an interaction, interactor, participant and experimental description, are sequentially generated numbers that are unique within a BIND translation build. The original BIND interaction or complex ID is saved as a primary reference within each entry.
Data ‘filtering’
The automated BIND translation process found and removed interaction entries that did not meet minimum requirements as defined by MIMIx. These entries were logged as PSI-MI interactions in a separate file that accompanies every translation build. ‘Filtered’ entries were defined as interactions having no primary reference for either one of its interactors (even if the interactor has a human readable name) and complexes referencing any ‘Filtered’ interaction.
CV mapping
BIND terms were mapped to their respective CV terms in the PSI-MI ontology for four data categories: interaction detection method, cellular localization, interactor type and xrefs (Supplementary Table S1). Interaction detection method, cellular localization and interactor type are all clearly defined in the BIND schema and a mapping for almost every term in the PSI-MI CVs or GO cellular component was found using the ontology look-up service (26). Since BIND does not have a CV for its xrefs, we manually translated them to PSI-MI CV terms. We also determined the reference type, and mapped these to corresponding PSI-MI CV terms (identity (MI:0356—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0356&termName=identical object)—reference to a corresponding object in another database; source reference (MI:0685—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0685&termName=source reference)—a publication reference describing where the interaction or curated information first appeared; primary reference (MI:0358—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0358&termName=primary-reference)—a publication reference describing the experimental data; see-also(MI:0361—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0361&termName=see-also)—reference to a related object in another database; or gene product (MI:0251—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0251&termName=gene product)—reference of a protein object to its corresponding genomic or nucleic acid sequence). For example, a UniProt (27) xref is considered an identity reference for proteins.
Identifier mapping and updating
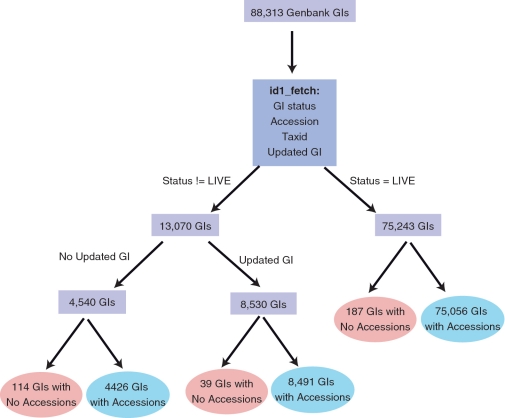
BIND preferred the use of Genbank GenInfo Identifiers (GIs) and Entrez Gene IDs (contained in the BIND-di field) to identify its protein, RNA, DNA and gene interactors. Since GIs tend to change often (any time a sequence changes), we mapped GIs to their corresponding accessions where possible (Figure 2) using an NCBI Toolkit command line tool id1_fetch (http://www.ncbi.nlm.nih.gov/IEB/ToolBox/CPP_DOC/lxr/source/src/app/id1_fetch/id1_fetch.cp). Accessions returned from id1_fetch originated from one of 7 databases including UniProt (27), RefSeq (28), EMBL (29), GenBank (30), PDB (31), DNA data bank of Japan (DDBJ) (32) or Protein information resource (PIR) (33). Accessions retrieved using id1_fetch were added as primary references in addition to the original GI that was stored as a secondary reference. All original GIs were annotated with their status as retrieved from id1_fetch. Status values could be Live, Live|Suppressed, Replaced, Replaced|Withdrawn, Replaced|Suppressed or Nonexistant. Updated GIs were added as secondary references with an attribute called ‘BIND translation id conversion’ with the value of ‘Updated GI’. NCBI Taxonomy Identifiers (Taxids) stored in BIND were updated to use the latest taxid retrieved through id1_fetch, if necessary.
Figure 2.
Identifier mapping process.
BIND records often contained references to additional databases, such as ChEBI for small molecules. All such references were translated to PSI-MI as secondary references but were neither updated nor validated against the original source database.
BIND PSI-MI validation
All output PSI-MI files were validated against the PSI-MI 2.5 schema using the official PSI-MI validator (http://www.ebi.ac.uk/intact/validator/start.xhtml), which checks a number of rules, including MIMIx compliance and correct use of CV terms (22).
Technologies used
All the source code for the BIND Translation process was written in Java (1.6), is open source, and is available at: http://baderlab.org/BINDTranslation. NetBeans (version 6.7), sponsored by Sun Microsystems (http://www.sun.com), was used as a Java IDE. JDOM (version 1.1) was used as an XML API and Xerces (version 2.0) as the XML parser, in addition to some use of the PSI-MI Java API. XML Spy (Professional Edition, version 2009, SP1) and Altova XML, both from Altova, Inc. (http://www.altova.com/) were used for visualizing/analyzing XML schemas, and for batch XML file validation, respectively.
Results
All BIND files were translated to PSI-MI 2.5 XML and MITAB and posted online at http://download.baderlab.org/BINDTranslation/. The translated BIND information provides verified and updated GIs, added accessions, error filtering and BIND CV to PSI-MI CV translation. The PSI-MI BIND repository has 206 859 unique interaction records, 88 313 identified unique interactors (based on their primary identifier), 3703 unique complexes and 16 643 unique PubMed references, involving 1512 identified species and 6 interactor types.
BIND element survey
The source BIND XML files validated, with minor errors, against the BIND XML schema. The minor errors were caused by some source BIND files, including experimental method types not defined in the BIND CV (‘microarray’ in Homo sapiens, 406 times and ‘synthetic-lethal-sick-test’ in Saccharomyces cerevisiae, 474 times).
Data issues and cleaning
BIND translation encountered several types of erroneous data. Table 3 shows some error categories with examples. These were either filtered (e.g. complexes referencing wrong interaction IDs were not translated) or automatically corrected, where possible (e.g. changing several ‘RefSeq’ xref types to ‘GI’). Mapped fields with missing or dummy data values in BIND (like ‘NULL’) were not translated if the PSI-MI schema did not require them. If required (e.g. interaction or interactor’s name), the corresponding PSI-MI elements were set to have a unified representation for the missing information (i.e. ‘NO_VALUE’) instead of the inconsistent formats BIND used to represent missing values.
Table 3.
Data cleaning: selected classes of errors, with examples, found in BIND
| Error type | Examples |
|---|---|
| No unified representation for missing information of type character/String | Missing information may be represented as: 'Unknown', 'NULL', 'unknown', ‘WP:NULL’, ‘unknown.’, ‘– ‘,…etc (in addition to ignoring the enclosing XML element altogether at times) |
| No unified representation for missing information of type integer | Missing information may be represented as: ‘0’, ‘–1’,…etc |
| Erroneous representation for references to external databases (x-ref) for some interactors | <BIND-other-db> |
| <BIND-other-db_dbname>LocusLink</BIND-other-db_dbname> | |
| <BIND-other-db_intp>0</BIND-other-db_intp> | |
| <BIND-other-db_strp>0</BIND-other-db_strp> | |
| </BIND-other-db> | |
| …. | |
| <BIND-other-db> | |
| <BIND-other-db_dbname>SGD</BIND-other-db_dbname> | |
| <BIND-other-db_intp>0</BIND-other-db_intp> | |
| <BIND-other-db_strp/> | |
| </BIND-other-db> | |
| Erroneous internal cross-reference: complexes referencing non-existent (negative) BIND interaction IDs | <BIND-mol-object-source_a> |
| Erroneous external cross-reference: negative PubMed identifier | PubMed ID ‘–2’ repeated 68 times in the S.Cerevisiae file |
| Inconsistent pattern for representing the IDs of some interactor x-refs | SGD identifiers ‘SGD: S000003663’ and ‘S000003663’; MGD identifiers ‘MGI:1890695’ and ‘1890695’ are all used. |
| Wrong x-ref type: listing some IDs as RefSeq identifiers while in fact they are GIs | GI IDs: ‘15643805’ and ‘15644490’ listed as RefSeq IDs. |
| Out dated external cross-references | There are 13 070 interactor GIs used in BIND that are not currently in use in Entrez. |
Identifier and CV mapping
From the total of 88 313 unique GIs, 87 973 GIs were successfully mapped to 61 106 unique accessions (Figure 2). Only 340 interactor GIs could not be mapped to accessions. A total of 75 243 GIs had ‘LIVE’ status. Of 13 070 GIs that were not marked as ‘LIVE’, 8530 GIs had an updated GI and 4540 did not have updated GIs. A total of 10 818 unique GIs had different taxonomy identifiers (taxids) specified in the BIND record as compared to those retrieved from id1_fetch and were updated. This was due to changes to the NCBI taxonomy classification that required new taxids to be created for those sequences. Also, we developed a one-to-one mapping between almost 400 BIND and PSI-MI or GO CV terms that were used by the translation process (Supplementary Table S1).
BIND species
Of 1587 species with an interactor in BIND, 74 taxids did not have a current match in Entrez Taxonomy, mostly due to species identifiers being renamed, merged or retired (Supplementary Table S2). This issue was fixed during identifier mapping by updating taxids using the ID Fetch system. BIND has inter-species interactions resulting in the same interaction being listed in two organism-specific source files. A list of duplicates (the duplicate BIND interaction ID and its matching PSI-MI interaction IDs) was generated so users can easily identify the duplicates when merging data from multiple files.
BIND PSI-MI validation
Our BIND PSI-MI files validated correctly against the PSI-MI schema, and for MIMIx standard compliance using the PSI-MI validator. There were a few errors relating to invalid taxids and our use of BIND CV identifiers for experimental method detection elements, which we were unable to translate due to the absence of equivalent CV terms in PSI-MI. These terms are candidates for adding to the PSI-MI CV (Table 4).
Table 4.
Proposed extensions to the PSI-MI ontology to allow CV mappings for currently unmatched BIND terms
| BIND term missing in PSI-MI CVs | Definition | Type of CV term | Web link |
|---|---|---|---|
| Beilstein | Compound database | Database citation (MI:0444)→participant database(MI:0473) | http://www.info.crossfiredatabases.com/home.shtml |
| EINECS | Compound database | Database citation (MI:0444) → participant database (MI:0473) | http://ecb.jrc.ec.europa.eu/esis/ |
| Merck | Compound database | Database citation (MI:0444) →participant database(MI:0473) | http://www.merckbooks.com/mindex/ |
| dictyBase | Organism Database | Database citation (MI:0444) →participant database (MI:0473) →sequence database (MI:0683) | http://dictybase.org/ |
| HGNC | Organism Database | Database citation (MI:0444) →participant database (MI:0473) →sequence database (MI:0683) | http://www.genenames.org/ |
| PlantGDB | Organism Database | Database citation (MI:0444) → participant database (MI:0473) → sequence database (MI:0683) | http://www.plantgdb.org/ |
| RatMap | Organism Database | Database citation(MI:0444) → participant database(MI:0473) → sequence database (MI:0683) | http://www.ratmap.org/ |
| TAIR | Organism Database | Database citation (MI:0444) → participant database (MI:0473) → sequence database (MI:0683) | http://www.arabidopsis.org/ |
| TIGR | Organism Database | Database citation(MI:0444) → participant database(MI:0473) → sequence database (MI:0683) | http://plantta.jcvi.org/ |
| ZFIN | Organism Database | Database citation (MI:0444) → participant database (MI:0473) → sequence database (MI:0683) | http://zfin.org/cgi-bin/webdriver?MIval=aa- ZDB_home.apg |
| COG | Protein Family Database | Database citation (MI:0444) → participant database (MI:0473) | http://www.ncbi.nlm.nih.gov/COG/ |
| Photon | Interactor type | ||
| Equilibrium dialysis | Method to detect interaction between Ligand and receptor under equilbrium conditions. | Interaction detection method (MI:0001) → experimental interaction detection(MI:0045) | |
| Membrane filtration | Method of filtration to separate molecules from a liquid | Participant detection method (MI:0002) → experimental participant identification (MI:0661) | |
| Monoclonal antibody-blockade | Method to block a binding site on a molecule, such as a protein, using a monoclonal antibody to test that the binding site is involved in an interaction with another molecule. | This term is probably too general to properly classify as an interaction detection method | |
| Nuclear translocation-assay | Method to detect interaction by inducing nuclear localization of one participant, which would then pull an interacting participant along with it into the nucleus. As both participants are labeled, the difference in nuclear localization between the induced and non-induced states provides an indication of the interaction between the two proteins. PMID: 20615205 | Interaction detection method (MI:0001) → experimental interaction detection (MI:0045) | |
| Transient-coexpression | Method consists of the expression of two proteins in a cell followed by interaction detection using a specific method. | This term is probably too general to properly classify as an interaction detection method | |
| Reconstitution | Method to reconstitute participants of a protein interaction in vitro to test if they bind. Depends on an interaction detection method | This term is probably too general to properly classify as an interaction detection method | |
| ASAP | No valid link available—Term Ignored | ||
| BMDL | No valid link available—Term Ignored | ||
| Locus tag | No valid link available—Term Ignored | ||
| MFCD | No valid link available—Term Ignored | ||
| MDL, MDL # | No valid link available—Term Ignored | ||
| aMAZE | Pathway database | Does not exist anymore |
Access via PSICQUIC web services and Cytoscape
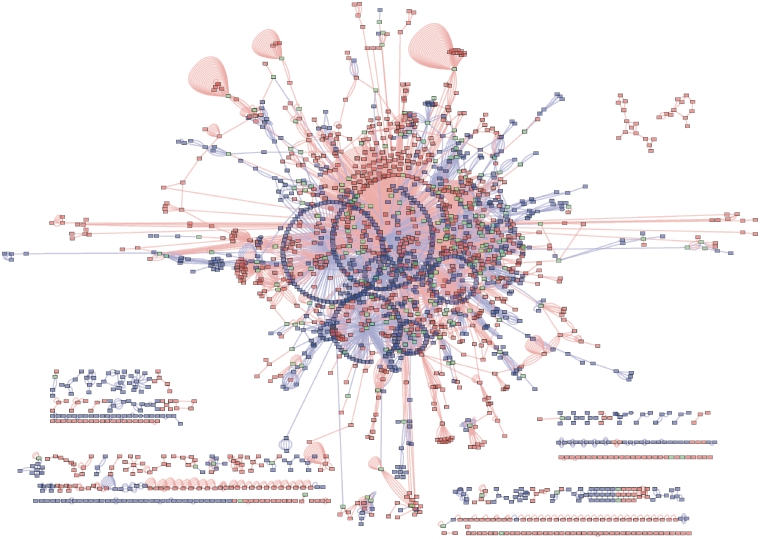
All interactions in BIND were imported into an instance of the Proteomics Standards Initiative Common Query Interface (PSICQUIC) interaction web service (http://webservice.baderlab.org:8180/psicquic-ws/). Figure 3 shows a network loaded into Cytoscape using the PSICQUIC plugin. The network consists of the union of all interactions from species Rattus norvegicus (taxid:10 116) from our current translation of BIND and IntAct. [PSICQUIC returns interactions for BIND (2738), BioGrid (632), DIP (92), IntAct (2629), MINT (2609), Matrix DB (40), iRefIndex (7414)]. IntAct was chosen because it contained the most interactions (besides iRefIndex) for this species. iRefIndex was not used for this analysis as it has already incorporated a prior build of the BINDtranslation and would not correctly reflect the added data that BIND offers. The BIND subset consists of 1103 unique nodes, and the IntAct subset consists of 984 unique nodes and they share 217 nodes. From this figure, we can clearly see the added data that BIND offers for this particular species.
Figure 3.
Union of BIND and IntAct interactions for species Rattus norvegicus (taxid: 10 116) as extracted using the PSICQUIC plugin for Cytoscape. Blue edges are interactions from IntAct, red edges are interactions from BIND. Blue nodes are interactors in IntAct only, red nodes are interactors in BIND only and green nodes are interactors shared by the two networks. BIND contains 1103 nodes not in IntAct. IntAct contain 984 nodes not in BIND. The two interaction networks share 217 nodes.
BIND PSI-MI download page
The resulting BIND data files in PSI-MI format can be downloaded from http://download.baderlab.org/BINDTranslation/. The current BIND translation build is labeled as release 1.0. All files are named by the taxid of the species they reference, one file per species. Users can download the ‘All species’ file or the ‘Selected model species’ file. These were selected based on their popularity or the number of interactions/complexes they contain. They hold ∼85% of all BIND interactions and complexes, and they are: Homo sapiens, Mus musculus, Rattus norvegicus, Saccharomyces cerevisiae, Drosophila melanogaster, Caenorhabditis elegans, Arabidopsis thaliana, Escherichia coli, Thermus thermophilus, Schizosaccharomyces pombe, HIV I, Escherichia coli, K-12, Helicobacter pylori (26 695), Bos taurus and Synthetic Construct interactions. Each build folder has its own log of ‘Filtered entries’.
Discussion
Our conversion of BIND to PSI-MI 2.5 XML and MITAB formats represents the most comprehensive coverage of BIND in the public domain, including complexes, all interactor types and species. The conversion, specifically the ID mapping, will be maintained over time. This makes BIND Translation the best public bulk download source of BIND MIs and complexes to date. The availability of BIND in PSI-MI 2.5 standard format and via the standard PSIQUIC web service enables the import of individual interactions, species sets or the entire database into research pipelines.
The BIND translation to PSI-MI involved major work in three areas: field mapping, CV term mapping and ID mapping. Since BIND was developed before the PSI-MI or other standard ontologies and CVs were developed by the community, BIND made use of its own data structures, CVs and preferred use of specific ID systems. Many of these were later standardized and best practices developed after BIND curation had stopped. The comparison of BIND best practices to current standards highlights specific design choices that were and were not successful in developing a lasting resource and provides a case study for interaction database design.
The BIND translation involved mapping fields between two different data models. This mapping is lossy if it fails to match elements with similar or identical meanings or if elements are present in one model and not in the other. In many cases, BIND fields perfectly matched PSI-MI fields (partially because BIND designers were involved in designing PSI-MI). BIND fields that could not be mapped to specific PSI-MI elements were mapped to general purpose elements in PSI-MI where possible (such as the generic attribute/value data type) or not mapped. For example, the PSI-MI XML schema does not have a record type for complexes, as BIND does, and instead models them as interactions with two or more participants or a set of interactions, where topology is known. As a result, BIND complexes were translated to a PSI-MI interaction, and the unsupported BIND complex attributes, including number of subunits and complex descriptions were mapped to generic PSI-MI attribute elements (clearly named and stored in interaction→attributeList). Importing systems can still automatically detect BIND complexes by customizing their PSI-MI file readers to read BIND translation-specific attributes. Attributes that were not mapped are discussed in future directions below.
On the other hand, some BIND entries did not contain the minimal information required for a PSI-MI entry, and artificial or unspecified values were used to ensure creation of valid PSI-MI records. For example, some BIND complexes reference a PubMed ID for a paper describing the complex, but had no interaction detection method information, which is required for adding a PubMed ID to a PSI-MI record (as a bibref element). To avoid losing these identifiers, we used the ‘unspecified (interaction detection) method’ CV term (MI:0686—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0686&termName=unspecified method) in all such entries. Also, every PSI-MI interaction requires a participant detection method, which is not captured in BIND. In this case, we used the ‘inferred by curator’ CV term (MI:0364—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0364&termName=inferred by curator) to comply with PSI-MI.
Apart from field mapping, a major amount of work was CV mapping. On the one hand, extensive use of CVs in BIND is extremely valuable for record consistency and ease of querying. On the other hand, BIND CVs were not adopted by the community, which led to some semantic mismatch converting BIND to PSI-MI CVs. BIND CVs were also not designed in a way that was easy to update, which led to their inconsistent use within the BIND database (see below for examples). Specifically, CVs were part of the BIND specification instead of an external dictionary. This increases the resources needed to update CVs as any update requires change of core software that validates and handles all records. Use of an external dictionary, as is done in PSI-MI, makes it easy to maintain evolving CVs without affecting the specification. Our mapping of these custom CVs to standard CVs, like GO and PSI-MI improves the ability to integrate BIND data with other sources and query the result.
We mapped three main BIND CVs to PSI-MI: interactor type (6 terms), interaction detection method (BIND-experimental-system: 42 terms), and cellular location (BIND-gen-place: 231 terms). However, the CV mapping between BIND and PSI-MI or GO is imperfect. Often, we could find a perfectly matched term, but sometimes we could only find a similar term or no term at all (Table 4). The CV for cellular locations in BIND was originally designed before GO, and its cellular component ontology, was available (34). Within this CV, we could map 95 terms to an identical GO cellular component, 128 terms to similar GO terms and 8 terms not at all, although no records contained terms that could not be mapped. As GO became more widely available and complete, it was used by BIND curators to describe cellular location (in BIND-spec-place). Although initially described as a human readable-specific text location in the BIND Curator manual, GO terms were eventually stored in this data structure, and these could easily be mapped to PSI-MI. When mapping CVs to the ‘closest possible’ match for a source term, in the absence of a perfect one, the available matching terms may be more general or specific than the source. In the absence of additional rules about the source term’s usage, the safer choice in this case is to select the more general matching term, since there is a guaranteed is–a relationship between the source and target terms. This was applied, for instance, when mapping both of the BIND interaction detection methods ‘allele-specific-complementation’ and ‘site-directed-mutagenesis’ as a PSI-MI ‘genetic interference (MI:0254—http://www.ebi.ac.uk/ontology-lookup/browse.do?ontName=MI&termId=MI:0254&termName=genetic interference)’. An example of a term that could not be mapped to PSI-MI is the BIND interactor type ‘photon’, which is used in 291 interactions in A. thaliana and S. cerevisiae. In case of an unmapped term, we translate it anyway so that the information is available, even if not standardized. For example, we use the term ‘photon’ for a PSI-MI interactor type, but do not reference a PSI-MI standard CV. Importing systems that do not support use of non-PSI-MI CVs will ignore these interactors or consider them as participants with an unknown type. Another example of unmapped terms exists in the xref type CV. There are many unmatched BIND xref terms in the PSI-MI ontology (Table 4). These are the less frequently used xrefs, many of which were used to reference non-protein interactors (e.g. Merck Index identifiers used for small molecule interactors). This is more of an indication of the diversity of BIND than a shortcoming of PSI-MI CVs. Thus, we have proposed potentially useful terms for incorporation in the PSI-MI CVs (4).
BIND lacks a CV for database names of interactor xrefs, but PSI-MI requires use of standard names for these. Unfortunately, BIND curators used different spellings when referencing databases. For example, the ecocyc pathway database name (http://ecocyc.org) was represented in three ways in BIND: ‘EcoCyc’, ‘ECOCYC’ and ‘EcoCyc ID’. This highlights the need to use standard terms for database names in any database, as implemented by the MIRIAM resource (35).
BIND relies on Entrez GIs as the primary identifier for sequence interactors (protein, DNA, RNA), with Entrez gene IDs taking a secondary role, though many other references to bioinformatics databases (xrefs) were used. Entrez GIs change with each new version of a sequence and are therefore generally less stable than the other identifiers, like Entrez gene IDs and UniProt or RefSeq protein accessions. The ID mapping process was successful at finding accessions for most (99.5%) BIND interactors. We did not add many additional IDs because a few standard IDs, especially for proteins and genes, is sufficient for users to gather other IDs from public ID mapping services themselves. As described in the ‘Results’ section, we were unable to map 340 interactor GIs to either updated GIs or accessions. This small percentage (<1%) was due to these GIs referencing sequence records that were permanently removed (versus being replaced) by NCBI, and are no longer traceable using the id1_fetch system.
One major issue resulting from the ID mapping process is the potential data compression as demonstrated by the smaller number of unique accessions that the unique GIs map to, i.e. multiple GIs map to the same accession. Examination reveals that 13 186 accessions (of 61 106) map to more than one GI (40 393 unique GIs total for the set). The remaining 47 920 accessions uniquely map to 1 GI. Accessions with multiple GIs all originate from the PDB database of protein structures, as a single structure could contain multiple molecules. Unfortunately, the NCBI ID Fetch system considers PDB IDs to be equivalent to sequence accessions, even though they are semantically different. By storing both the PDB accession and associated GI, we retain a link to the original sequence referenced in the interaction and this enables a future conversion of these to correct sequence accessions.
In order to transfer binding site information in the future, we need to translate the position on the original sequence to the updated sequence or make sure that ID mapping adds only identifiers representing the exact same sequence. The sequence of the original record must be compared to any new sequence references we add for a correct mapping. Our identifier mapping process shows that 75 243 GIs are LIVE, thus binding sites can be transferred directly. For the remaining 13 070 GIs, the original sequences need to be compared to the updated sequences in order to transfer binding site annotation.
The ID mapping process and the retired species identifiers we found (‘Results’ section) show how BIND became partially out of sync with major bioinformatics data warehouses, resulting in significant loss of curated information over only ∼4 years. For example, BIND used the taxid 11 489 (Influenza A virus) that has been changed to 132 504 in Entrez. Cross-references that break over time will require updating in future translation operations, but using more standard identifier types, such as accessions, will likely improve the longevity of the data. It is important that database providers work harder to prevent this slow degradation by using better systems to track identifier updates over time and update their records accordingly. This is especially important for MI and pathway databases, which make use of a large amount of cross-references to molecular databases.
The BIND specification was designed based on a biochemical paradigm and was highly detailed so as to capture as much molecular biology as possible (from interaction to atomic level detail). However, our element survey shows that most (3070 of the total 3542 elements, or 87%) of these fields were used in less than a quarter of the interactions. Thus, the approach by PSI-MI to focus on mature, frequently used data types results in a much higher percentage of specification use. On the other hand, using only a limited number of fields can not capture all knowledge in the literature. This highlights the continued need to improve data models and develop standard abstractions in biology.
While BIND is currently owned by Thomson Reuters, Inc., the BIND data we translated is available in the public domain, which means anyone can use and redistribute it without restriction. We do not plan to create new BIND records, but only maintain a translation that helps users access the data in a standard format and with current identifiers. Thomson Reuters created a commercial version of BIND called BINDPlus, which contains over 180 000 additional, mostly automatically derived or high-throughput interactions, with some manually curated interactions. Both BIND and BINDPlus are now static resources since early 2009, thus we can consider the BIND data set a stable, but still useful and unique resource.
Future directions
Our aim in this project was to provide a reliable and complete translation for the most frequently used BIND interaction components for the entire BIND repository. We also refined and updated the parts of BIND that we translated and made it available to the scientific community. As shown by our XML element survey, there are other, less frequently used, BIND fields that were not covered by our translation. These include interaction binding action, binding states and binding sites. We hope to cover these in a future version of the translator. Binding sites can be mapped to PSI-MI participant features, but actions and states are not covered by PSI-MI—though they are covered by the BioPAX pathway exchange language (36). We also hope to further refine our translation, by increasing coverage for publication references in interactions and inferring interaction types from experimental methods.
Supplementary Data
Supplementary data are available at Database Online.
Funding
U.S. NIH via National Human Genome Research Institute (NHGRI) (grant P41 P41HG04118); Genome Canada through the Ontario Genomics Institute (2007-OGI-TD-05). Funding for open access charge: NIH grant (P41 P41HG04118).
Conflict of interest. None declared.
References
- 1.Alfarano C, Andrade CE, Anthony K, et al. The Biomolecular Interaction Network Database and related tools 2005 update. Nucleic Acids Res. 2005;33:D418–D424. doi: 10.1093/nar/gki051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader GD, Betel D, Hogue CW. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bader GD, Donaldson I, Wolting C, et al. BIND–The Biomolecular Interaction Network Database. Nucleic Acids Res. 2001;29:242–245. doi: 10.1093/nar/29.1.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hogue CW. The other side of staying out of a BIND. Nat. Biotechnol. 2007;25:971. doi: 10.1038/nbt0907-971a. [DOI] [PubMed] [Google Scholar]
- 5.Breitkreutz BJ, Stark C, Reguly T, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aranda B, Achuthan P, Alam-Faruque Y, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JY, Mamidipalli S, Huan T. HAPPI: an online database of comprehensive human annotated and predicted protein interactions. BMC Genomics. 2009;10(Suppl 1):S16. doi: 10.1186/1471-2164-10-S1-S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Han K, Park B, Kim H, et al. HPID: the Human Protein Interaction Database. Bioinformatics. 2004;20:2466–2470. doi: 10.1093/bioinformatics/bth253. [DOI] [PubMed] [Google Scholar]
- 10.Chaurasia G, Malhotra S, Russ J, et al. UniHI 4: new tools for query, analysis and visualization of the human protein-protein interactome. Nucleic Acids Res. 2009;37:D657–D660. doi: 10.1093/nar/gkn841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ng A, Bursteinas B, Gao Q, et al. pSTIING: a ‘systems’ approach towards integrating signalling pathways, interaction and transcriptional regulatory networks in inflammation and cancer. Nucleic Acids Res. 2006;34:D527–D534. doi: 10.1093/nar/gkj044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lynn DJ, Winsor GL, Chan C, et al. InnateDB: facilitating systems-level analyses of the mammalian innate immune response. Mol. Syst. Biol. 2008;4:218. doi: 10.1038/msb.2008.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Razick S, Magklaras G, Donaldson IM. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prieto C, De Las Rivas J. APID: Agile Protein Interaction DataAnalyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tarcea VG, Weymouth T, Ade A, et al. Michigan molecular interactions r2: from interacting proteins to pathways. Nucleic Acids Res. 2009;37:D642–D646. doi: 10.1093/nar/gkn722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jensen LJ, Kuhn M, Stark M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McDowall MD, Scott MS, Barton GJ. PIPs: human protein-protein interaction prediction database. Nucleic Acids Res. 2009;37:D651–D656. doi: 10.1093/nar/gkn870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brown KR, Jurisica I. Unequal evolutionary conservation of human protein interactions in interologous networks. Genome Biol. 2007;8:R95. doi: 10.1186/gb-2007-8-5-r95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salama JJ, Donaldson I, Hogue CW. Automatic annotation of BIND molecular interactions from three-dimensional structures. Biopolymers. 2001;61:111–120. doi: 10.1002/1097-0282(2001/2002)61:2<111::AID-BIP10143>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 21.Hermjakob H, Montecchi-Palazzi L, Bader G, et al. The HUPO PSI's molecular interaction format–a community standard for the representation of protein interaction data. Nat. Biotechnol. 2004;22:177–183. doi: 10.1038/nbt926. [DOI] [PubMed] [Google Scholar]
- 22.Montecchi-Palazzi L, Kerrien S, Reisinger F, et al. The PSI semantic validator: a framework to check MIAPE compliance of proteomics data. Proteomics. 2009;9:5112–5119. doi: 10.1002/pmic.200900189. [DOI] [PubMed] [Google Scholar]
- 23.Bader GD, Hogue CW. BIND–a data specification for storing and describing biomolecular interactions, molecular complexes and pathways. Bioinformatics. 2000;16:465–477. doi: 10.1093/bioinformatics/16.5.465. [DOI] [PubMed] [Google Scholar]
- 24.Orchard S, Salwinski L, Kerrien S, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 25.Bader GD, Hogue CW. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 26.Cote RG, Jones P, Martens L, et al. The Ontology Lookup Service: more data and better tools for controlled vocabulary queries. Nucleic Acids Res. 2008;36:W372–W376. doi: 10.1093/nar/gkn252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Apweiler R, Maria Jesus M, O’Donovan C, et al. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kulikova T, Akhtar R, Aldebert P, et al. EMBL Nucleotide Sequence Database in 2006. Nucleic Acids Res. 2007;35:D16–D20. doi: 10.1093/nar/gkl913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Karsch-Mizrachi I, Ouellette BF. The GenBank sequence database. Methods Biochem. Anal. 2001;43:45–63. [PubMed] [Google Scholar]
- 31.Kirchmair J, Markt P, Distinto S, et al. The Protein Data Bank (PDB), its related services and software tools as key components for in silico guided drug discovery. J. Med. Chem. 2008;51:7021–7040. doi: 10.1021/jm8005977. [DOI] [PubMed] [Google Scholar]
- 32.Satoru M. [Deposition with DNA Date Bank of Japan (DDBJ); its data format and tools for submitions] Tanpakushitsu Kakusan Koso. 2002;47:733–736. [PubMed] [Google Scholar]
- 33.Wu C, Nebert DW. Update on genome completion and annotations: protein Information Resource. Hum. Genomics. 2004;1:229–233. doi: 10.1186/1479-7364-1-3-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ashburner M, Ball CA, Blake JA, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laibe C, Le Novere N. MIRIAM Resources: tools to generate and resolve robust cross-references in Systems Biology. BMC Syst. Biol. 2007;1:58. doi: 10.1186/1752-0509-1-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demir E, Cary MP, Paley S, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]