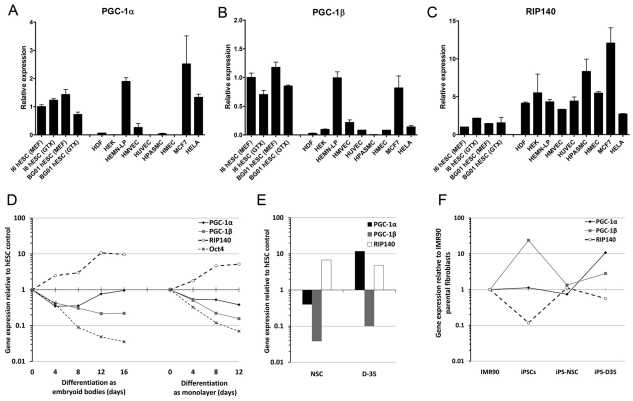
Fig. 3.
Gene expression profiles for three metabolic master regulators highlight similarities between pluripotent stem cells and other energy demanding cell types. Real-time reverse transcription–PCR data for expression of the mRNA encoding PGC-1α (A), PGC-1β (B) and RIP140 (C) in I6 and BG01 hESC lines maintained on feeder cells (MEF) or on Geltrex in conditioned medium (GTX) compared with seven other primary cell types and two cancer cell lines. HDF, human diploid fibroblast; HEK, human epidermal keratinocyte; HEMN-LP, human epidermal melanocyte-low pigmented; HMVEC, human mammary vein endothelial cells; HUVEC, human umbilical vein endothelial cells; HPASMC, human pulmonary artery smooth muscle cells; HMEC, human mammary epithelial cells, MCF7, breast cancer cell line; and HeLa, cervical cancer cell line. Results represent means+s.e.m. for 2–4 independent RNA preparations. (D) Real-time reverse transcription–PCR data during differentiation of hESCs as embryoid bodies or as a monolayer. Data represent the mean values for two hESC lines (H9 and I6). (E) mRNA expression for NSCs and D-35 cells relative to their parental hESC lines. (F) mRNA expression changes during reprogramming of IMR31 fibroblasts into iPSCs and differentiation into iPSC-NSCs and iPSC-D35 cells.