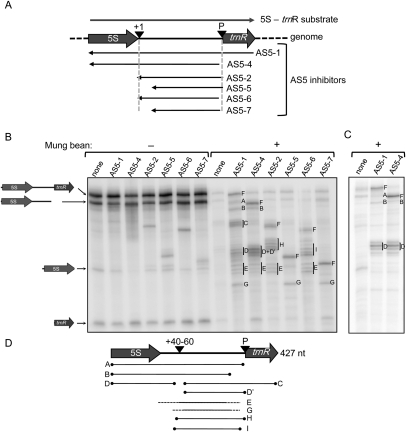
FIGURE 7.
Mapping intermolecular base-pairing between 5S-trnR and AS5 by treatment with mung bean nuclease. (A) Representation of the substrates used for the experiment in panels B and C. (+10) An endonuclease cleavage site; (P) RNAse P cleavage site. (B) Labeled 5S-trnR was incubated with the chloroplast protein extract in duplicate with or without a 500-fold excess of each AS5 inhibitor. Reaction products were treated (+) or not (−) with mung bean nuclease following RNA purification, and analyzed by 5% denaturing polyacrylamide gel electrophoresis. (A–I) Transcripts referred to in the text and in panel D, with F representing the putative full-length sense-antisense duplex for each AS5 inhibitor. Where clusters of bands occur, a vertical line marks the bands assigned to that cluster. (C) 5S-trnR was 5′-end-labeled and incubated with chloroplast protein extract with or without a 500-fold excess of the two inhibitors shown. RNA was extracted prior to treatment with MBN. (D) Interpretation of bands seen in panels B and C, as discussed in the text. (+40–60) The region of discontinuity susceptible to MBN digestion.