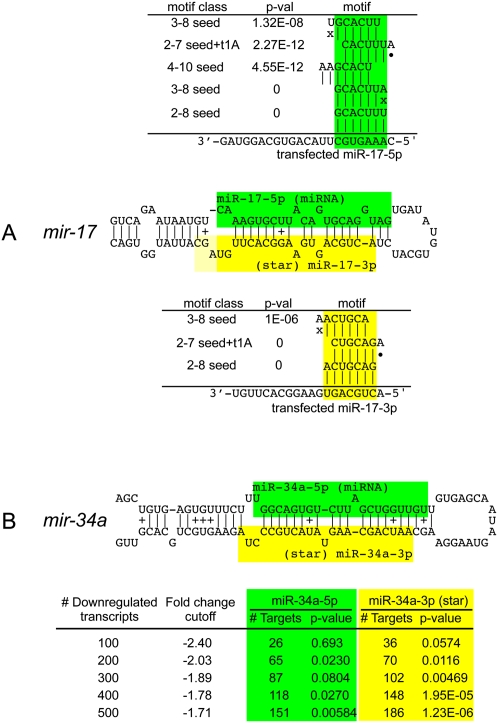
FIGURE 4.
Transcriptome-wide evidence for target repression directed by partner miRNA and star species. (A) Target repression by mature (green) and star (yellow) products of mir-17. Linsley et al. (2007) reported microarray profiles for cells transfected with mimics for either miR-17-5p (mature) or miR-17-3p (star), but only analyzed the mature strand response. Using miREDUCE, we observe that the motifs that are most highly correlated with down-regulated transcripts in these data sets are the miR-17-5p seed (2–8 or 3–8) and the miR-17-3p seed (2–8 or 2–7seed + t1A), respectively, all with P-values = 0. Other highly statistically enriched motifs in down-regulated transcripts include various seed variants (see also Supplemental Table 2). (B) Mendell and colleagues reported microarray profiles following infection with a retroviral mir-34a construct (Chang et al. 2007) and reported that the miR-34a seed was enriched among the 100 most down-regulated transcripts. miREDUCE on this transcript set yielded statistically significant enrichment of miR-34a 2-7 seed (green) among the top 200 down-regulated transcripts, but this enrichment increased when larger sets of down-regulated transcripts were analyzed. These same transcript subsets yielded even stronger enrichment for miR-34a* seeds (yellow) across all comparable cohorts of down-regulated genes, peaking at 1.23E-06 in the top 500 most down-regulated transcripts.