FIGURE 5.
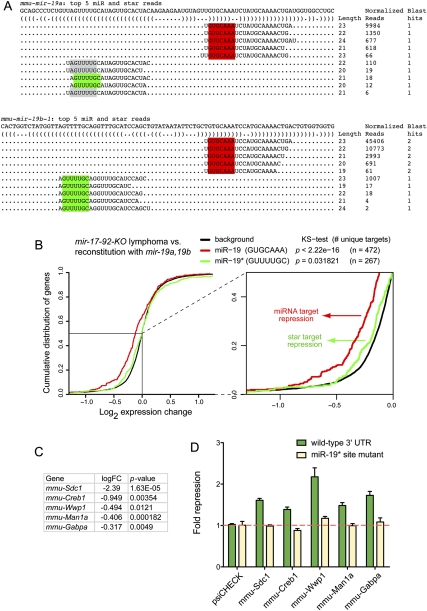
Evidence for targeting by endogenous miR-19 and miR-19*. (A) Dominant read counts of mmu-mir-19a and mmu-mir-19b-1 analyzed from six data sets reported in GSE11724 (Marson et al. 2008; see also Supplemental Fig. 2). Both genes generate a mature species with a shared seed (red box); miR-19a* generates two 5′ isomiRs, one of which is shared with miR-19b-1* (green box); note that the GUUUUGC miR-19* seed is by far the dominant star seed generated by the mir-19 genes. (B) Cumulative distribution function of gene expression in lymphoma cell lines deleted for the mir-17→92 cluster compared to those re-expressing mir-19a/b under retroviral control. Transcripts whose 3′ UTRs contained exclusively miR-19 (GUGCAAA) or miR-19* (GUUUUGC) seed sites are segregated for analysis; transcripts containing both types of seed matches were discarded to avoid ambiguity in assigning the targeting species. Transcripts with miR-19 seed sites (red) or miR-19* seed sites (green) were down-regulated in the presence of mir-19 relative to background gene expression of all other transcripts (black); this trend was more substantial for the mature strand miR-19 but still statistically significant for star strand targets. The region boxed in the main graph is expanded to illustrate this trend more clearly. (C) Fold change of transcript down-regulation for selected targets bearing conserved miR-19* seed matches. Following multiple hypothesis correction by the false discovery rate (FDR), log fold changes with adjusted P-values (FDR <%5) were considered significant; all of these gene expression changes were highly significant. (D) Confirmation of direct repression by miR-19*. 3′-UTR target sensors and matched mutants bearing specific point changes within the miR-19* seed matches were tested for their response to transfection of mir-19a/b in HeLa cells; sensor activities were normalized to their level in the presence of functional mir-1-2 construct. All five were repressed in a manner that was completely dependent on the integrity of the miR-19* seed matches (cf. wild-type sensors in green with mutant sensors in tan).