Figure 6. Leucine catabolism in bacteria and humans.
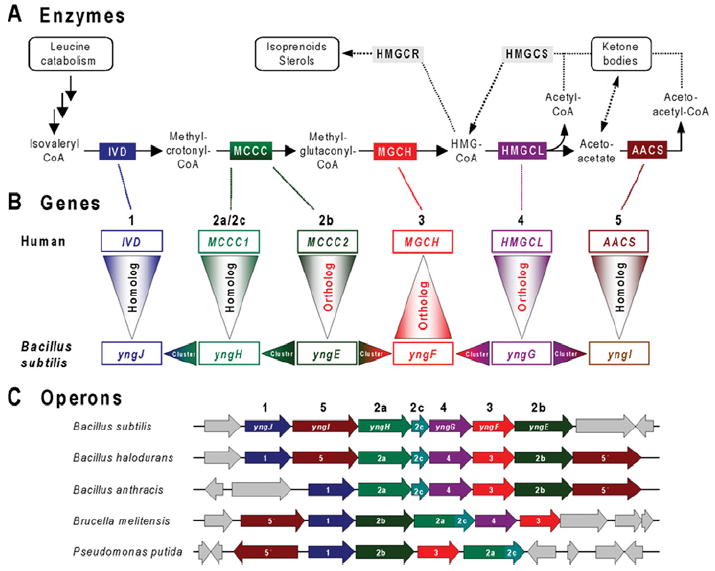
(A) Enzymatic steps involved in the later steps of leucine catabolism and the metabolism of hydroxymethylglutaryl-CoA (HMG-CoA). Intermediates are shown by chemical names. Enzymes conserved in humans, Bacillus subtilis and certain other bacteria are as follows: IVD, isovaleryl-CoA dehydrogenase (EC 1.3.99.10); MCCC, methylcrotonoyl-CoA carboxylase (EC 6.4.1.4) [2a/2c, biotin-carboxylase subunit/biotin carboxyl carrier domain/subunit; 2b, carboxyl transferase subunit]; MGCH, methylglutaconyl-CoA hydratase (EC 4.2.1.18); HMGCL, hydroxymethylglutaryl-CoA lyase (EC 4.1.3.4); and AACS, acetoacetate-CoA synthetase (EC 6.2.1.16). These five enzymes are colour-coded. Two enzymes related to the mevalonate pathway of isoprenoid/sterol biosynthesis (present in humans, but not B. subtilis) are shown in grey: HMGCS, HMG-CoA synthase (EC 2.3.3.10); and HMGCR, HMG-CoA reductase (EC 1.1.1.34). Enzymes catalysing early steps of leucine catabolism (from leucine to isovaleryl-CoA) are similar in humans and bacteria (not shown). (B) Projection of functional assignments between human and bacterial genes. Gene names (human and B. subtilis) corresponding to pathway enzymes are colour-coded and numbered in agreement with (A). The reasoning used in analysing leucine catabolism in bacteria is illustrated by arrowheads pointing in the direction of functional projections. Vertical triangles with red lettering correspond to unambiguous projections based on orthology (same specific function). The triangles point in the direction of the projection. Vertical triangles with black lettering indicate homologues that belong to large families that contain multiple paralogues that share a ‘general class’ function, but differ in substrate specificity. Horizontal triangles indicate conjectures based on gene clustering (refinement of ‘general class’ functions and genuine functional predictions). (C) Large operon-like clusters of genes related to leucine catabolism detected in a number of Gram-positive and Gram-negative bacteria. Conserved homologous genes are colour-coded and numbered in agreement with (A) and (B). Genes without homologues in a given chromosomal neighbourhood are coloured grey.
