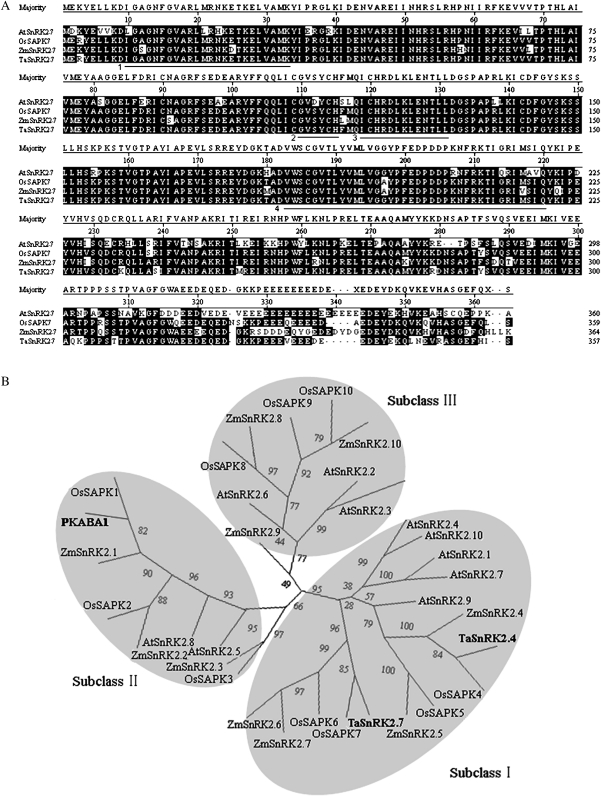
Fig. 1.
Sequence alignment of TaSnRK2.7 and SnRK2s from other plant species. (A) Alignment of the predicted amino acid sequences of TaSnRK2.7 and closely related plant SnRK2 protein kinases. Conserved prosite motifs are underlined. Regions 1–4 represent the ATP binding site, N-myristoylation site, protein kinase activating signature, and transmembrane-spanning region, respectively. Alignments were performed using the Megalign program of DNAStar. Common identical AARs are shown with a black background. Dashed lines represent gaps introduced to maximize alignment. Abbreviations on the left of each sequence: Ta, T. aestivum; Os, O. sativa; At, A. thaliana; Zm, Z. mays. (B) Phylogenetic tree of TaSnRK2.7 and SnRK2s from other plant species. Three distinct isoform groups are presented in grey. The phylogenetic tree was constructed with the PHYLIP 3.68 package; bootstrap values are in percentages.