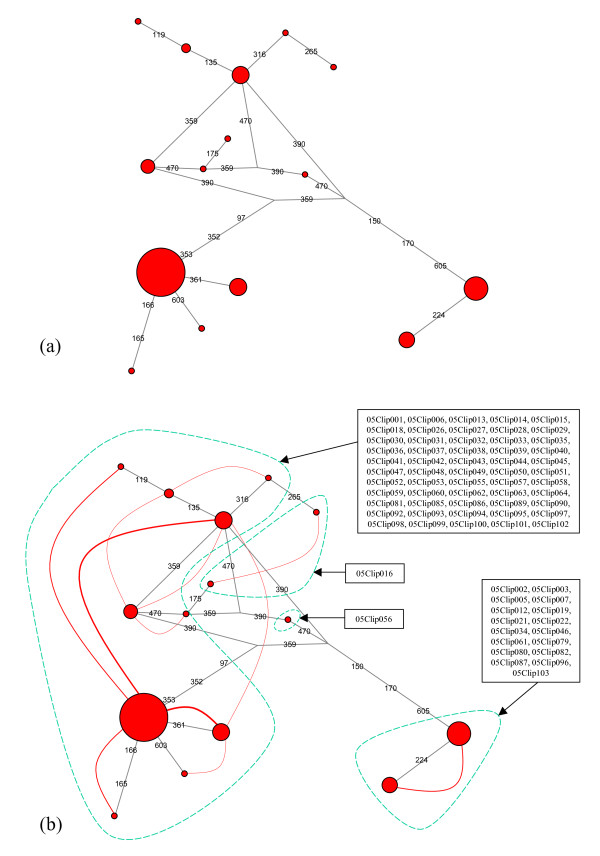
Figure 3.
ITS haplonet and haploweb. (a) As a first step in the analysis, a haplotype network (in short, "haplonet") was build from the alignment of all ITS sequences. Since 31 individuals were heterozygous and 43 were homozygous for this marker, the alignment comprised 105 sequences, among which 15 different haplotypes could be distinguished (represented as circles on the haplonet, with diameters proportional to the number of individuals harboring each of them). Each haplotype is connected to one or several others by straight lines representing the evolutionary paths inferred by the network-building algorithm (numbers in red on the lines represent mutated positions in the alignment). (b) As a second step, the haplonet was converted into a haplotype web (in short, "haploweb") by adding curves connecting haplotypes found co-occurring in heterozygous individuals (the width of each curve is drawn proportional to the number of heterozygotes harboring the two haplotypes it connects). This allowed us to delineate 4 pools of co-occurring haplotypes (enclosed in green dashes on the figure). To each of these 4 allele pools corresponds a group of individuals (called single-locus fields for recombination, or sl-FFR in Doyle's terminology), whose names are listed inside boxes with arrows pointing on the corresponding allele pool: these 4 sl-FFRs comprised respectively 55, 17, 1 and 1 individuals.