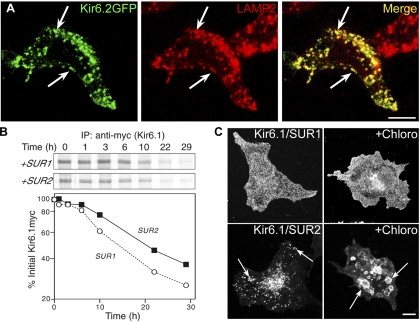
Fig. 2.
Kir6.2/SUR2A channels localize to endosomes and lysosomes, but this does not affect the protein turnover rate. A: HEK-293T cells transfected with Kir6.2-green fluorescent protein (GFP)/SUR2A cDNAs were stained for IFM with rat anti-human lysosomal-associated membrane protein-2 (LAMP2) and rhodamine-conjugated anti-rat antibody. There was extensive overlap between the GFP signal and the staining for the lysosome/late endosome marker, LAMP2 (arrows). B: HEK-293T cells transfected with plasmids encoding Kir6.1myc and either SUR1 or SUR2A were subjected to pulse-chase labeling. After immunoprecipitation (IP), samples were subjected to SDS-PAGE (B, top), and the Kir6.1myc band was quantified and plotted as a function of the chase time (B, bottom). Depicted is 1 of 3 similar experiments. C: COS-1L cells transfected with Kir6.1myc and SUR1 or SUR2A cDNAs were pretreated with chloroquine (Chloro; 100 μM) and cycloheximide for 5 h before staining for IFM with anti-myc antibodies. The drug caused the intracellular vesicles to enlarge (arrows). Figure panels have been adjusted for brightness and contrast. Bars = 10 μm.