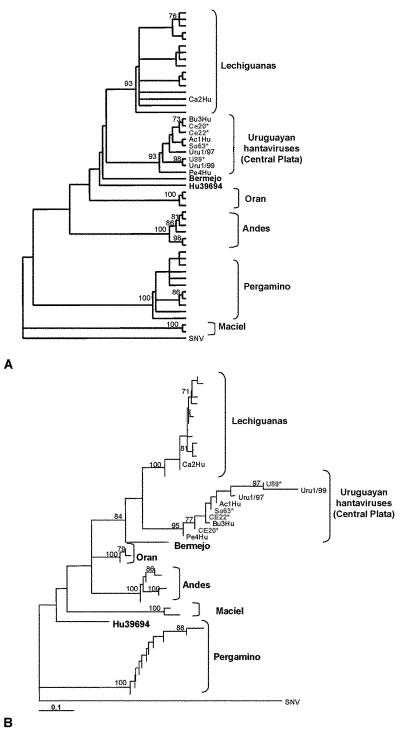
Figure 2.
A: Maximum parsimony phylogenetic tree B: Distance-based phylogenetic tree. The tree was built under the Tamura-Nei model of DNA substitution with estimation of the shape parameter of the gamma distribution ( 28 ). This model and the associated parameters resulted from testing our dataset with the program MODELTEST 3.06 ( 26 ). Both trees include Argentinean and Uruguayan hantaviruses from hantavirus pulmonary syndrome (HPS) case-patients and rodents. Hantavirus sequences from HPS case-patients in Uruguay: Ac1Hu, Ca2Hu, Bu3Hu, Pe4Hu, Uru1/97, Uru1/99. Hantavirus sequences from yellow pygmy rice rats: U89, Sa63, Ce20, Ce22. Sin Nombre virus was used as outgroup.*Specimens deposited at the Specimen Collection of the Sección Zoología Vertebrados, Facultad de Ciencias, Universidad de la República, Montevideo, Uruguay, with the following numbers: U89=ZVC-M2154, SA63=ZVC-M2155, Ce20=ZVC-M2156, Ce22=ZVC-M2157, and Ce155=ZVC-M2158.