Abstract
Previous genetic studies in Sphingomonas macrogolitabida strain TFA have established that expression of genes involved in tetralin biodegradation (thn genes) requires the function of the LysR type activator ThnR and also ThnY. Sequence comparison indicated that ThnY is homologous to bacterial oxygenase-coupled NAD(P)H-dependent ferredoxin reductases. However, ThnY showed substitutions in highly conserved positions of the pyridine nucleotide binding domain of these ferredoxin reductases. ThnY expression is co-regulated with all other genes required for tetralin biodegradation, and presumably thnY is part of the thnCA3A4RY operon. ThnY has been purified, and its biochemical and functional properties were characterized. ThnY was found to be a monomeric orange-brown iron-sulfur flavoprotein (estimated mass of 37,000 Da) containing one non-covalently attached flavin adenine dinucleotide and one plant type ferredoxin 2Fe-2S cluster. It can be efficiently reduced by dithionite, but reduction by pyridine nucleotides was very poor. Consistently, ThnY-dependent reduction of cytochrome c, ferricyanide, or 2,6-dichlorophenolindophenol using NAD(P)H as the electron donor was undetectable or very weak. The addition of ThnY to electrophoretic mobility shift assays containing ThnR and a probe bearing two thn divergent promoters resulted in a 3-fold increase in protein-DNA complex formation affinity, which indicates that ThnY directly promotes thn transcription activation by ThnR.
Keywords: Bacterial Signal Transduction, Electron Transfer, FAD, Flavoproteins, Gene Expression, Gene Regulation, NADH, Reductase, Bacteria Biodegradation, Dioxygenases
Introduction
Tetralin (1,2,3,4-tetrahydronaphthalene) is a bicyclic molecule composed of an aromatic and an alicyclic moiety, which is found at low concentrations in different crude oils, and it is also industrially produced for its use as an organic solvent. Biodegradation of tetralin has been characterized most extensively in Sphingomonas macrogolitabida strain TFA. The genes required for tetralin biodegradation (thn genes) have been sequenced, and the functions of the encoded proteins have been characterized to elucidate the biodegradation pathway (1–4) that yields pyruvate and pimelate, which is subsequently metabolized via β-oxidation (5). Rieske type oxygenases are multicomponent enzymes known to initiate the oxidative biodegradation of various environmentally hazardous aromatic compounds in bacteria, such as crude oil components, polycyclic aromatic hydrocarbons, and heterocyclic aromatic compounds, in a reaction requiring oxygen and an external electron supply (6). In this reaction, the electrons are transferred from NAD(P)H via flavin and [2Fe-2S] redox centers to the terminal oxygenase component that has the catalytic active site. Tetralin dioxygenase is encoded by the thnA1 and thnA2 genes, whereas the ferredoxin and the ferredoxin reductase components of the electron transfer system are encoded by thnA3 and thnA4, respectively. According to the number of components and the nature of the redox centers of the electron transport chain, bacterial oxygenases have been classified into three main classes, classes I, II, and III (7–9). The tetralin dioxygenase system belongs to class III (4).
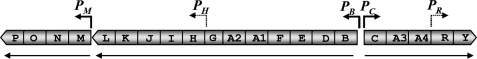
The thn genes involved in utilization of tetralin as a carbon and energy source by Sphingomonas macrogolitabida strain TFA cluster together and are arranged into three closely linked operons, two of which contain internal promoters (Fig. 1). With the exception of the weak constitutive thnR promoter, transcription from the thn promoters is co-regulated at a transcriptional level in a complex regulatory manner. First, thn expression is specifically induced in the presence of tetralin but prevented if other preferential carbon sources are available, implying that additional signals, such as the physiological status of the cell or catabolite repression, might be integrated into the regulatory system (5, 10). Second, oxidation and reduction of the ferredoxin component ThnA3 apparently act as a molecular switch to control thn gene expression. This conclusion is based on the observation that under circumstances where the transfer of electrons from the ferredoxin to the tetralin dioxygenase is blocked (i.e. in the absence of a suitable substrate for the dioxygenase or in a mutant lacking tetralin dioxygenase), reduced ThnA3 would accumulate, and thn gene expression would be prevented. It has been proposed that this system will avoid gratuitous induction by potential inducers that are not substrates of the terminal oxygenase (11).
FIGURE 1.
Transcriptional organization of the thn genes. thn genes are organized in a 14.3-kb cluster. Genes are marked by boxes, with the direction of the arrowheads indicating the direction of transcription. The arrows below the genes indicate the three main transcriptional units. The bent arrows indicate the identified transcription initiation sites and the direction of transcription. Internal promoters are marked with dashed arrows. Transcription from the weak PR promoter is constitutive, whereas transcription from the others is regulated by tetralin.
Transcription from the regulated thn promoters requires a positive control by regulatory proteins. Previous genetic analysis showed that S. macrogolitabida mutant strains defective in either thnR or thnY were unable to activate thn gene transcription (5, 10). ThnR is a LysR type of transcriptional activator whose regulatory mechanism starts to emerge. Several lines of evidence, including semiquantitative and quantitative expression analysis by RT-PCR of several thn genes, footprint analysis, and DNA binding assays, have shown that transcription from the PB, PC, PH, and PM promoters is positively regulated by ThnR in response to tetralin (5, 12). It has recently been shown that ThnR binds to palindromic sites present at each of the four thn promoter regions in order to activate transcription. Like most LysR type activators, ThnR apparently binds as a tetramer to contiguous high affinity and low affinity sites in each promoter region. Additionally, a mechanism of co-regulation of the transcription from the closely located divergent thnB and thnC promoters has been reported, which involves the formation of a DNA loop and a higher order structure maintained by interaction between ThnR molecules bound to each promoter region (12), thus generating an octameric protein structure. As shown in Fig. 1, thnR is located downstream of the structural thnC, thnA3, and thnA4 genes. Expression analysis by quantitative RT-PCR has shown that thnR has a low constitutive transcription level from its own promoter, but its transcription is also induced by tetralin from the thnC promoter (12).
However, unlike ThnR, ThnY does not show homology to any reported transcriptional regulator. In fact, ThnY shows a clear homology to oxidoreductases that participate as electron transfer components to terminal oxygenases. Apart from its essential requirement for in vivo thn gene transcription, nothing is known about the role and the physiological partners of ThnY. It would be interesting to know why and how a protein similar to ferredoxin reductases is essential for thn gene transcription. In this report, we have begun to investigate the function of ThnY. We present here the purification of ThnY and some of its molecular, biochemical, and functional properties and propose a regulatory role of ThnY as modulator of the activity of the transcriptional activator ThnR.
EXPERIMENTAL PROCEDURES
Media and Growth Conditions
Escherichia coli strains were routinely grown in Luria-Bertani (LB) medium at 37 °C (13). S. macrogolitabida strains were grown at 30 °C in mineral medium plus 2 g of tryptone liter−1 and 1 g of yeast extract liter−1 (MML)-rich medium (2) or MM medium (14) supplemented with either 8 or 40 mm β-hydroxybutyrate (βHB)2 as the carbon source. When required, tetralin was provided in the gas phase (15). Antibiotics were used, when required, at the following concentrations: ampicillin (100 μg/ml or 5 μg/ml for E. coli or S. macrogolitabida strains, respectively), kanamycin (20 μg/ml), gentamicin (10 μg/ml), and chloramphenicol (15 μg/ml).
Construction of Strains and Plasmids
Bacterial strains and plasmids used in this work are listed in Table 1.
TABLE 1.
Bacterial strains and plasmids used in this work
| Bacterial strains and plasmids | Relevant characteristics | Reference/Source |
|---|---|---|
| Strains | ||
| E. coli DH5α | F− φ80d lacZΔM15 Δ(lacZYA–argF)U169 recA1 endA1 hsdR17 (rk−mk−) supE44 thi-1 gyrA relA1 | Ref. 49 |
| E. coli NCM631 | hsdS gal λDE3:lacI lacUV5:gen1(T7 RNA-polymerase) Δlac linked to Tn10 | Ref. 50 |
| S. macrogolitabida TFA | Wild type. Strr | Ref. 15 |
| S. macrogolitabida T601 | thnY::mini-Tn5Km, inserted in PstI, codon 140. Strr, Kmr | Ref. 10 |
| S. macrogolitabida T601–1002 | T601 with a thnC-lacZ translational fusion into the chromosome. Strr, Apr, Kmr | Ref. 10 |
| S. macrogolitabida T1032 | 780-bp thnR in-frame deletion. Strr | Ref. 12 |
| S. macrogolitabida T1032–1002 | T1032 with thnC-lacZ translational fusion into the chromosome. Strr, Apr | Ref. 12 |
| Plasmids | ||
| pET23b | Expression vector. Apr | Novagen |
| pIZ227 | pACYC184 containing lacIq and the pLysE. Cmr | Ref. 50 |
| pMPO759 | Expression vector containing thnY in pET23b, for overproduction of ThnY-His6. Apr | This work |
| pIZ1020 | pT7-thnR, with His6 tag fused in frame to C terminus. AUG initiation codon. Apr | Ref. 12 |
| pIZ1016 | pBBRMCS-5 broad host range vector derivative, with the tac promoter and laclq. Gmr | Ref. 10 |
| pIZ698 | thnY in pIZ1016. Gmr | Ref. 10 |
| pMPO750 | thnY in pIZ1016. Gmr | This work |
| pMPO761 | ThnY-His6 expressed from the ptac promoter in pIZ1016. Gmr | This work |
| pMPO513 | 269-bp fragment in pBluescript II SK+, bearing the wild type thnB–thnC intergenic region. Apr | Ref. 12 |
Plasmid pMPO750 harboring the thnY gene was obtained by insertion of a 0.97-kb BamHI-EagI DNA fragment from pIZ698 into the broad host range expression vector pIZ1016 digested in the same way, resulting in thnY gene expression from the tac promoter. The ThnY-His6-overproducing plasmid pMPO759 was constructed amplifying thnY by PCR with primers NdeI-thnY (5′-GGCATATGGAAATCACCCTCATC-3′) and thnY-XhoI (5′-GGCTCGAGCGAAACAGAAAAATG-3′), using pMPO750 as the template. The PCR product was cleaved with NdeI and XhoI and cloned into pET23b digested with the same enzymes. To test the functionality of the ThnY-His6, the plasmid pMPO761 carrying that gene expressed from a tac promoter was constructed by insertion of a Bpu1102I (Klenow)-XbaI DNA fragment from pMPO759 into the broad host range expression vector pIZ1016 digested with SacI (Klenow) plus XbaI. For complementation analysis, plasmids pMPO750 and pMPO761 were transferred into recipient strains T601–1002, and transconjugant colonies were selected on MML agar plates by using resistance to gentamicin, ampicillin, kanamycin, and streptomycin.
E. coli DH5α was used as a host in all cloning procedures. All DNA manipulations were performed according to standard procedures (13). Plasmid DNA was transferred to E. coli and S. macrogolitabida strains by transformation or by triparental mating, respectively.
Induction Assays
Induction assays with tetralin were performed as described previously (10); briefly, S. macrogolitabida TFA-1002 (wild type) and T601-1002 (thnY miniTn5km) strains and the same strains complemented with the indicated plasmids were grown at 30 °C in MM medium containing 40 mm βHB as the only carbon and energy source to the exponential phase (optical density at 600 nm of 0.8) and the corresponding antibiotics. Then cells were washed to remove the carbon source and diluted to a final A600 of about 0.1 in MM medium with antibiotics, supplemented with 8 mm βHB in the absence or presence of the tetralin. Tetralin was supplied in the gas phase. Cultures were grown at 30 °C for 22 h, and β-galactosidase activity was assayed as described by Miller (16).
Western Blotting
Crude cell extracts of different S. macrogolitabida TFA strains were prepared from cells grown under carbon-rich (40 mm βHB) or carbon-limited (8 mm βHB) conditions either in the presence or absence of tetralin as indicated above. Proteins were run on 12% (w/v) SDS-PAGE. For Western blotting, polyclonal antiserum against S. macrogolitabida ThnY raised in rabbit were used, and the antibody-antigen reaction was visualized using the chemiluminescent method (Thermo Scientific) as recommended by the manufacturer.
Expression and Purification of Recombinant ThnY-His6
Cultures of E. coli NCM631 harboring the plasmid pIZ227 and the ferredoxin reductase thnY expression plasmid pMPO759 were grown in LB medium containing 100 μg/ml ampicillin and 15 μg/ml chloramphenicol overnight. They were used to inoculate 1 liter of Terrific Broth medium (13) plus the corresponding antibiotics and incubated at 37 °C until A600 reached 0.3. At this point, cells were incubated for 30 min at 30 °C. Expression from the φ10 promoter from the T7 phage was induced by adding 0.5 mm isopropyl β-d-thiogalactopyranoside. Extra iron and sulfur sources (0.1 mg/ml ferric ammonium citrate, 0.1 mg/ml ferric citrate, 0.1 mg/ml iron sulfate heptahydrate, 0.1× ferrous sulfate/chelate solution, and 1 mm cysteine) were added to the culture medium. The incubation was continued for 15 h at 16 °C. Cells were harvested by centrifugation at 4000 × g for 20 min at 4 °C, and the cell pellet was stored at −80 °C until further use.
His-tagged ThnY was purified using the commercial cobalt affinity chromatography IMAC column (TALONTM resin (Clontech)) according to the manufacturer's specifications. All of the following procedures were carried out at 4 °C. Cell pellets were resuspended in the lysis buffer (50 mm Tris-HCl buffer, pH 7.4, containing 250 mm NaCl, 10% glycerol) and 1 mm phenylmethylsulfonyl fluoride and disrupted by sonication. Broken cells were ultracentrifuged at 90,000 × g for 30 min, and the supernatant was loaded onto an IMAC column pre-equilibrated with the lysis buffer containing 10 mm imidazole. The column was washed extensively with the lysis buffer, and bound proteins were eluted with an increasing imidazole gradient. Fractions containing the protein were collected and concentrated. Size exclusion chromatography was performed with a Superdex 75 pg column (10/60) using the lysis buffer with a constant flow rate of 0.2 ml/min. Suitable fractions were collected, concentrated, and stored at −80 °C. The apparent molecular mass of ThnY was determined by calibrating the column with a commercial filtration calibration kit for low molecular weight proteins (GE Healthcare) according to the manufacturer's specifications. Protein concentration was calculated using the BC assay kit (Uptima/Interchim) according to the manufacturer's protocol, and purity was estimated visually by SDS-PAGE to be about 95%.
ThnR-His6 Purification
ThnR purification was performed as described previously with some modifications (12). Cells were chilled, harvested by centrifugation, and mechanically lysed using Grinder (6870 Freezer/Mill). Cells were broken with five cycles using the following program: 10-min precool, 2-min run, 2-min cool (14 counts/s). Broken cells were resuspended in 150 ml of chilled purification buffer (20 mm sodium phosphate, pH 7.4, 500 mm NaCl, 10 mm imidazole, 5% glycerol) and ultracentifuged at 90,000 × g, and the supernatant was collected to ThnR purification. Imidazole (75 mm) was used to elute the protein, which was subsequently stored at 80 °C.
UV-visible Spectroscopy
UV and visible absorption spectra of the protein from 250 to 800 nm were measured using a Shimadzu UVPC-1603 double beam spectrophotometer. Time course measurements of the reduction of ThnY were performed in the lysis buffer by adding an excess of either dithionite (0.6 mm) or NAD(P)H (20 mm) as reducing agents in comparison with the protein concentration (35 μm). Anaerobic reduction was performed in an anaerobic chamber, and the spectrophotometric measurements were carried out keeping the anaerobic conditions. All of the buffers were flushed with argon for more than 2 h to remove all of the oxygen. UV-visible spectra of the oxidized or reduced forms of ThnY were recorded in 50 mm Tris-HCl buffer, pH 7.4, containing 250 mm NaCl, 10% glycerol.
Flavin Determination
Purified ThnY was boiled in the dark for 10 min and centrifuged to remove the denatured protein. The cofactor of the protein was visualized by resolving the supernatant at room temperature and in the dark by TLC on silica sheets Alugram SilG (20 × 20 cm, thickness 0.20 mm) plates. The mobile phase was a solution of butanol/acetic acid/water (12:3:5). Determination of flavin TLC spots was done by exposing the TLC plate under UV light (17). A solution of commercial FAD (50 μm) and FMN (50 μm) was used as a standard. After identification of the flavin as FAD, its concentration was quantified spectrophotometrically using the extinction coefficient ϵ450 nm = 11,300 m−1 cm−1.
EPR Spectroscopy
EPR spectra were recorded on a Bruker ESP300 spectrometer (X-band, 9.5 GHz) equipped with a continuous flow helium cryostat ESR 900 and an ITC 4 temperature controller (Oxford Instruments) to allow measurements down to 5 K. Parameters were as follows: modulation frequency, 100 kHz; microwave power, 2 milliwatts; modulation amplitude, 5 G; time constant, 20 ms, temperature, 20 K. The microwave frequency was measured by an HP 5350B frequency counter. Because flavins can only be detected in EPR in the semiquinone form and also [2Fe-2S] clusters are EPR-silent in the oxidized state, the ThnY samples (150 μm) were reduced with either dithionite (0.6 mm) or NADPH (20 mm) before they were transferred into EPR quartz tubes and frozen in liquid nitrogen. To obtain absorption spectra of the reduced [2Fe-2S] centers, the samples (150 μm) in oxidized form or after reduction with dithionite (0.6 mm) or NAD(P)H (20 mm) were transferred in EPR quartz tubes and frozen in liquid nitrogen.
Enzymatic Assays
Enzymatic activity of ThnY was measured by reduction of cytochrome c (ϵ550 nm = 21 mm−1 cm−1), ferricyanide (ϵ420 nm = 1.02 mm−1 cm−1), and 2,6-dichlorophenolindophenol (DCPIP), sodium salt (ϵ620 nm = 21 mm−1 cm−1). Cytochrome c reduction assays were performed at 25 °C in 50 mm phosphate buffer, pH 7.4, containing 200 μm horse heart cytochrome c and different concentrations of ThnY. The reaction was initiated by the addition of NAD(P)H (200 μm), and the increase in A550 was followed spectrophotometrically over 10 min. Potassium ferricyanide and DCPIP reduction assays were performed in the same way except that 100 μm K3Fe(CN)6 or 100 μm DCPIP was used in place of cytochrome c. The reaction was monitored spectrophotometrically recording absorbance at 420 or 620 nm, respectively. One unit of activity was defined as the amount of the enzyme required to reduce 1 μmol of electron acceptor/min mg protein.
EMSAs
DNA probes of the thnB-thnC intergenic region (269 bp) containing both putative ThnR binding sites were obtained as SalI-EcoRV fragments of pMPO513. The probes were [α-32P]dCTP-labeled by Klenow filling of the 3′ recessed ends. EMSAs were performed as described previously (18). ThnR-DNA complexes were generated in 15 μl of reactions containing 2 fmol of each probe, 100 ng of salmon sperm DNA, 5 mg of BSA, and different amounts of purified ThnR-His6 and ThnY-His6. Gels were dried and exposed on a radiosensitive screen. Bands were visualized with a Typhoon 9410 scanner and analyzed using the ImageQuant software (GE Healthcare). The apparent dissociation constant (Kd) was calculated as the concentration of protein at which 50% of the probe was in the complex (19).
RESULTS
thnY Expression Pattern
As shown in Fig. 1, the thn genes, whose products are involved in aerobic biodegradation of tetralin by S. macrogolitabida strain TFA, are clustered in 14.3 kb and transcribed from three main and two internal promoters (5, 10, 12). Previous genetic analyses have demonstrated that transcription of thn operons is specifically induced by tetralin and repressed if other alternative carbon sources are present in the medium. Furthermore, several lines of evidence, including semiquantitative and quantitative expression analysis by RT-PCR of several thn genes, footprint analysis, and DNA binding assays, have shown that transcription from the PB, PC, PH, and PM promoters is positively regulated by ThnR in response to tetralin (5, 12).
The thnY gene is located downstream of the thnC, thnA3, thnA4, and thnR genes, which have recently been shown to be part of the same transcriptional unit (12). Moreover, the translational start codon of ThnY is only 7 bp downstream of the stop codon of the regulatory ThnR protein. These data suggest that thnY may be part of the same transcriptional unit that would then comprise five genes.
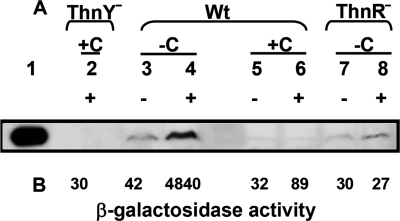
Because ThnY is indispensable for expression of the thn genes and as a first step in understanding the role of ThnY, we have determined the conditions necessary for synthesis of ThnY. To do that, cell extracts of strain TFA grown with 8 mm β-hydroxybutyrate (carbon-limiting conditions) or 40 mm β-hydroxybutyrate (carbon-rich conditions) with and without tetralin were used for Western blot experiments using ThnY antibodies as described under “Experimental Procedures.” Fig. 2A shows the levels of ThnY in the wild type and thnR mutant strains under different growth conditions. As a control, thnC expression, measured as the levels of β-galactosidase activity obtained from the thnC-lacZ gene fusions integrated into the chromosomes of the wild type and mutant thnR strains growing under the same growth conditions, is also shown (Fig. 2B). In the wild type strain, ThnY synthesis was induced under carbon-limited conditions when tetralin was present in the medium. However, under carbon-rich conditions, ThnY could not be detected regardless of the presence of tetralin. As mentioned above, transcription of the thn operons is under the control of the transcriptional activator ThnR. ThnY production is detectable but cannot be induced in the mutant strain lacking the thnR regulatory gene even under inducing conditions (Fig. 2A, lanes 7 and 8). These results indicate that synthesis of ThnY requires both tetralin and carbon-limited conditions and that this specific induction is dependent on ThnR, like the expression pattern described for the other thn genes.
FIGURE 2.
Western blot analysis showing levels of ThnY in crude cell extracts of S. macrogolitabida wild type and mutant strains under different growth conditions. A, plus and minus signs indicate the presence or absence of the inducer tetralin, respectively; +C and −C indicate carbon-rich and carbon-limited conditions, respectively. Lane 1, purified ThnY; lane 2, thnY miniTn5-km grown with tetralin and 8 mm βHB (induction conditions); lanes 3 and 4, WT strain grown on 8 mm βHB (carbon-limited conditions); lanes 5 and 6, WT strain grown on 40 mm βHB (catabolite repression conditions); lanes 7 and 8, in frame ΔthnR mutant grown on 8 mm βHB. B, expression of thnC-lacZ translational fusions integrated into the chromosome of each strain measured as β-galactosidase activity in Miller units.
Sequence Analysis of the S. macrogolitabida thnY Gene Product
ThnY encodes a 324-amino acid polypeptide, and its entire deduced amino acid sequence showed similarity to ferredoxin reductase components of the electron transport chain that transfer reducing equivalents from NAD(P)H to the catalytic site of terminal oxygenases. Bacterial oxygenases are enzymes that catalyze the insertion of molecular oxygen into aromatic benzene rings and play an important role in the degradation of aromatic compounds. Batie et al. (7) grouped the multicomponent oxygenase systems into classes I–III based on the number of protein components and on the nature of the redox centers of the electron transport chain. Class I is a two-component system that consists of a reductase and a terminal dioxygenase. Class I reductase components contains flavin (FMN in class IA and FAD in class IB) and a chloroplast type [2Fe-2S] center. Class II enzymes are three-component dioxygenase systems in which the flavin (FAD) and the [2Fe-2S] center (IIA, chloroplast type; IIB, Rieske type) are located on separate components. Class III comprises three-component systems, whose reductases harbor both FAD and a chloroplast type [2Fe-2S] cluster and transfer electrons to an intermediary Rieske type ferredoxin rather than directly to the oxygenase component. The overall degrees of sequence identity and similarity of ThnY were 36 and 56% to ferredoxin reductases of class III dioxygenase systems and 22 and 43% to ferredoxin reductases of the class IB branch, respectively.
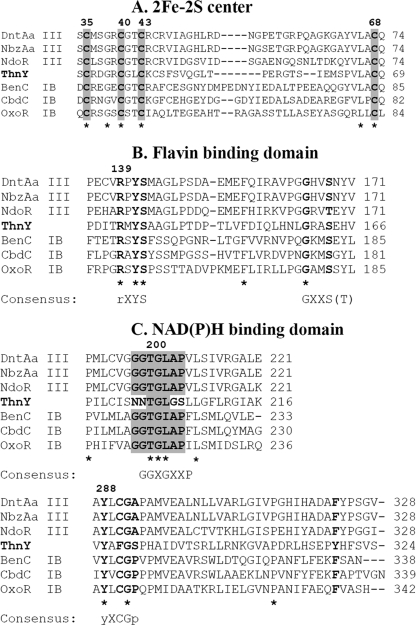
Classes IB and III share a common three-domain structure with structural determinants for binding of three redox cofactors. Fig. 3 shows the sequence alignment of the three ThnY domains with those of some ferredoxin reductases representative of these classes of enzymes (i.e. the well characterized BenC, the reductase for the benzoate 1,2-dioxygenase type IB system from Acinetobacter sp. ADP1, whose crystal structure has been reported (20), and the well known example of the type III enzyme of the naphthalene dioxygenase complex from Pseudomonas sp. NCIB 9816–4 (NdoR) (21)). All members of these reductases share an N-terminal ferredoxin domain containing the conserved residues Cys-X4-Cys-X2-Cys-X29/30-Cys for plant type [2Fe-2S] cluster binding. The central and C-terminal domains contain motifs that have been proposed to be involved in flavin and NAD(P)H binding, respectively, which closely resembles the plant and bacterial ferredoxin:NAD(P)+ reductase (FNR) superfamily of flavoproteins (22, 23). In fact, this two-domain module forms a unit often referred as the “FNR-like module.” A pattern of conserved residues located in the flavin and pyridine nucleotide-binding region proposed to be the characteristic signature of the FNR family (24) is shown in Fig. 3.
FIGURE 3.
ThnY sequence alignment. Amino acid sequence alignment of the three cofactor binding motifs of class IB and III reductases of ring hydroxylating oxygenases is shown. The class designation is based according to the system of Batie et al. (7). The asterisks indicate residues conserved in the six sequences, whereas dots indicate residues conserved in at least four sequences. A, ferredoxin domain; conserved cysteines for the plant type [2Fe-2S] cluster are shaded and in boldface type. B, FAD binding domain; conserved residues for binding of isoalloxazine and phosphate groups of the FAD, respectively, are shown in boldface type. C, NAD(P)H binding domain; glycine fingerprints that are highly conserved among all members of the plant/bacterial type FNRs and are involved NAD(P)H binding are shaded, and other conserved amino acids and the aromatic residue involved in the interaction with the adenine moiety of FAD are also indicated in boldface type. DntAa, 2,4-dinitrotoluene dioxygenase reductase from Burkholderia sp.; NbzAa, nitrobenzene dioxygenase reductase from Comamonas sp.; NdoR, naphthalene dioxygenase reductase from Pseudomonas sp.; BenC, benzoate 1,2-dioxygenase reductase from Acinetobacter sp.; CbdC, 2-halobenzoate 1,2-dioxygenase reductase from Pseudomonas cepacia; OxoR, 2-oxo-1,2-dihidroquinoline 8-monooxygenase reductive from Pseudomonas putida.
ThnY contains the iron-sulfur cluster domain and the flavin binding domain with the same features and arrangements conserved in both classes of reductases, including the consensus sequence for binding of isoalloxazine and phosphate groups of the FAD, respectively. However, all attempts to reveal a proper NAD binding domain failed. Although the alignments showed that some conserved features of NAD binding domains can still be recognized in ThnY, the highly conserved glycine signature (GGXGXXP) is not present because the two first glycines are replaced by two asparagines and the conserved proline is replaced by serine. Furthermore, the conserved cysteine corresponding to the consensus motif (yXCGp, where lowercase residues are less well conserved) proposed to be involved in binding of the nicotinamide group is also replaced by a phenylalanine. This residue and the proline from the sequence GGXGXXP have been proposed to contact the bound nicotinamide in the model of phathalate dioxygenase reductase complexed with NADH (25). Thus, sequence analysis indicated that ThnY might be involved in redox reactions but should not be reduced directly by pyridine nucleotides.
In Vivo Properties and Purification of ThnY-His6
In order to purify the protein in substantial amounts, ThnY was modified by the addition of six histidine residues at the C terminus. Prior to the purification, we considered the possibility that the addition of the His residues had some effect on the folding and on the activity of the protein. Thus, we first tested whether the His-tagged version of the protein is as active in vivo as the wild type protein (Table 2). As previously reported (10), ThnY is strictly required for expression of tetralin biodegradation genes, so a mutant lacking ThnY (T601), was unable to grow with tetralin as a sole source of carbon and energy. The mutant T601, which bears a thnC::lacZ translational fusion into the chromosome as a reporter of thn gene expression, was transformed with the isopropyl β-d-thiogalactopyranoside-inducible expression vectors pMPO750, bearing thnY, and pMPO761, bearing thnY-His6. Both plasmids complemented the thnY mutation because they allowed the mutant strain to grow on tetralin as the only carbon and energy source and restored the levels of thn expression to the wild type levels when tetralin was added into the medium (Table 2). A similar pattern of expression was obtained when isopropyl β-d-thiogalactopyranoside was added to the cultures (data not shown), which indicated that basal expression levels from these plasmids are enough to produce an amount of ThnY sufficient to complement the chromosomal mutation. Thus, there was no apparent difference in physiological activity between both forms of ThnY, and therefore ThnY-His6 can be used for subsequent in vitro studies.
TABLE 2.
Expression of the thnC-lacZ translational fusion in a thnY mutant strain and complementation with the His-tagged thnY gene
All of the strains were grown in mineral medium containing 8 mm β-hydroxybutyrate (carbon-limited conditions) as described under “Experimental Procedures.”
| Strain | β-Galactosidase activity |
|
|---|---|---|
| Without tetralin | With tetralin | |
| Miller units | ||
| TFA-1002 (wild type) | 42 ± 10 | 4844 ± 581 |
| TFA-1002 (pIZ1016) | 37 ± 10 | 3397 ± 969 |
| T601-1002 (thnY miniTn5km) | 29 ± 5 | 30 ± 5 |
| T601-1002/pMPO750 (Ptac-thnY) | 26 ± 6 | 2758 ± 343 |
| T601-1002/pMPO761 (Ptac-thnY-His6) | 27 ± 7 | 2661 ± 466 |
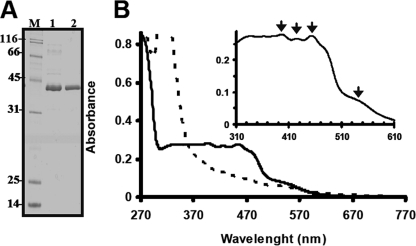
ThnY-His6 was overproduced in the E. coli strain NCM631 transformed with the plasmid pMPO759, which transcribes thnY-His6 from the φ10 promoter from the T7 phage. Optimal production of the enzyme was obtained by growing transformed cells at 16 °C for 15 h after isopropyl β-d-thiogalactopyranoside induction. After purification using a two-step procedure, as described under “Experimental Procedures,” inspection of a SDS-polyacrylamide gel showed a 95% pure protein of ∼37 kDa (Fig. 4A). The molecular mass of the purified protein was confirmed to be 37 kDa by gel filtration chromatography. This experimentally determined value is in good agreement with that of 35 kDa calculated from its coding sequence. Hence, ThnY seems to be a monomer and has an Mr almost identical to those its related counterparts.
FIGURE 4.
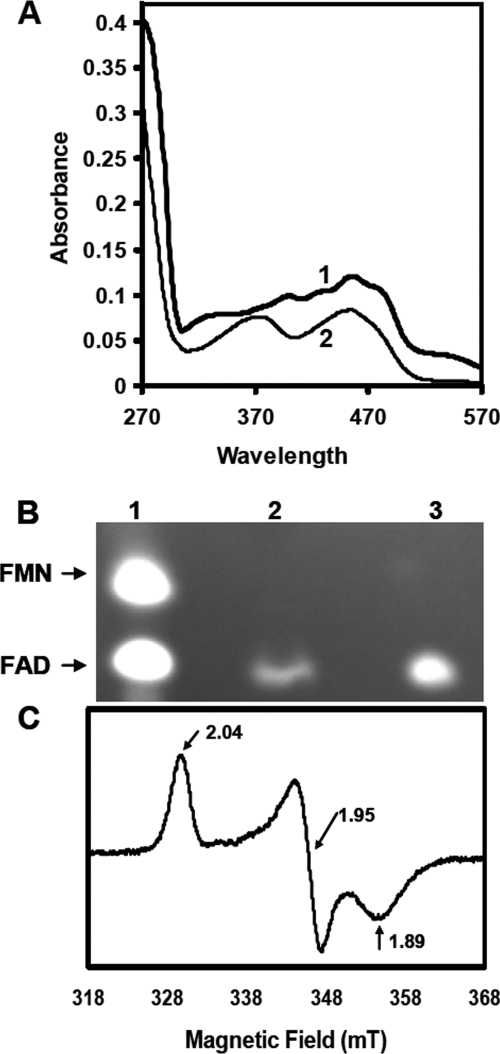
Purified ThnY and UV-visible absorption spectra. A, SDS-PAGE of purified ThnY; M, molecular weight marker; lane 1, purified ThnY after cobalt affinity chromatography; lane 2, purified ThnY after gel filtration chromatography. B, UV-visible absorption spectra; solid line, oxidized spectrum of ThnY as isolated; dashed line, spectrum after the addition of sodium dithionite. The protein concentration was 35 μm in 50 mm Tris-HCl buffer, pH 7.4, containing 250 mm NaCl, 10% glycerol. The inset shows a detail of the oxidized absorption spectrum between 310 and 610 nm.
ThnY Is an Iron-Sulfur Flavoprotein with a [2Fe-2S] Cluster and FAD as the Prosthetic Group
Preparations for protein purification and purified protein were orange-brown in color, indicative that some chromophores must be bound to the protein. The UV-visible spectrum of ThnY-His6 (Fig. 4B, solid lines; see inset for details) revealed absorption maxima at 270, 395, 420, and 453 nm and a shoulder at about 540 nm in the air-oxidized state. This spectrum is consistent with the sum of the spectra of bound oxidized flavin (maxima at about 370 and 450 nm) (26) and an oxidized [2Fe-2S] center (maxima at about 320, 415, and 455 nm) (27). The nature of these centers was therefore investigated and identified by biochemical methods and spectroscopic analyses. Similar spectral features have been reported for those reductases containing flavin and a plant type [2Fe-2S] cluster that function in transferring electrons from NAD(P)H to the dioxygenases systems and appear to be structurally related (e.g. benzoate 1,2-dioxygenase reductase shows absorption maxima at 273, 340, 402, and 467 nm (28), napthalene dioxygenase reductase at 278, 340, 420, and 460 nm (shoulder at 540 nm) (29), 2-halobenzoate 1,2-dioxygenase reductase at 272, 343, and 457 nm (30), and carbazole dioxygenase reductase at 278, 390, and 461 nm (31)). The purified ThnY was bleached under aerobic conditions upon the addition of an excess of sodium dithionite, as shown by the general decrease in the absorbance over the visible range (Fig. 4B, dashed lines), which clearly indicated that the cofactors could be reduced.
In most flavoproteins, the cofactor is strongly but non-covalently bound. This seems to be the case for ThnY, because a bright yellow compound was released from the purified protein after denaturation by boiling the sample for 10 min. The absorption spectrum of this chromophore exhibited maxima at 375 and 450 nm in the visible region, typical of flavin species (Fig. 5A, curve 2). Moreover, the flavin was identified as FAD by comparison of the sample with commercial FAD and FMN using thin layer chromatography, as described under “Experimental Procedures” (Fig. 5B). The amount of the FAD in the purified protein was calculated by measuring the extinction at 450 nm. Based on the molar extinction coefficient of FAD at 450 nm (ϵ = 11,300 m−1 cm−1) and the calculated extinction coefficient of the FAD of ThnY (19.318 m−1 cm−1), the molar ratios of FAD/protein determined from different preparations were 1.05 ± 0.15. These results suggest that 1 mol of ThnY contains 1 mol of FAD and that not much flavin is lost during the course of purification. This is also in agreement with the constant A270/A450 ratio found in the different preparations despite reports of loss of the flavin cofactor in similar proteins, such as the reductase component of naphthalene dioxygenase (29).
FIGURE 5.
Identification of the redox centers of ThnY. A, curve 2 shows the absorption spectra of the flavin extracted from ThnY compared with the wild type protein (curve 1). B, Identification of the flavin as FAD by TLC. Determination of flavin TLC spots was done by exposing the TLC plate under UV light as indicated under “Experimental Procedures.” Lane 1, a commercial solution containing 50 μm FMN and 50 μm FAD that were used as standards; lane 2, supernatant obtained after heat treatment of ThnY; lane 3, a sample of commercial FAD. C, EPR spectra of reduced ThnY. Purified ThnY (150 μm) was reduced aerobically with an excess of sodium dithionite. Measurement conditions were as follows: modulation frequency, 100 kHz; microwave power, 2 milliwatts; modulation amplitude, 5 G; time constant, 20 ms; temperature, 20 K.
Although the UV-visible spectra of vertebrate type and plant type ferredoxins are more or less identical, the two types of 2Fe-2S clusters are easily distinguishable by EPR. Therefore, we performed EPR measurements in order to further characterize the 2Fe-2S center of ThnY. The dithionite-reduced ThnY displays a rhombic EPR spectrum (Fig. 5C), with apparent g factors of g1 = 2.04, g2 = 1.95, and g3 = 1.89. The obtained values agree very well with those of typical plant type ferredoxins, such as spinach ferredoxin (g1 = 2.05, g2 = 1.96, and g3 = 1.89) or parsley ferredoxin (g1 = 2.05, g2 = 1.96, and g3 = 1.90) (32). Thus, the preliminary classification of the ThnY 2Fe-2S cluster as plant type on account of the conserved motif Cys-X4-Cys-X2-Cys-X29/30-Cys could be conclusively confirmed by EPR measurements. Hence, our preparations of purified ThnY contain all of the cofactors characteristics expected from its deduced amino acid sequence.
Reduction of ThnY by NAD(P)H
It is clear from these results that ThnY carries two redox centers that can be reduced with sodium dithionite. Ferredoxin reductases that are involved in electron transfer from NAD(P)H to terminal oxygenases are known to have NAD(P)H-dependent diaphorase activity and cytochrome c reductase activity; in these reactions, electrons are transferred from NAD(P)H to various artificial electron acceptors, such as DCPIP, ferricyanide, and cytochrome c. The activity of these enzymes varies according to the specificity they have for both coenzymes, NADH and NAD(P)H. ThnY did not support measurable cytochrome c reduction with either NADH or NADPH as reductant, and when we used DCPIP as an external electron acceptor, the levels of activity were extremely low, 7 × 10−4 μmol/min/mg of protein, regardless of the coenzyme used.
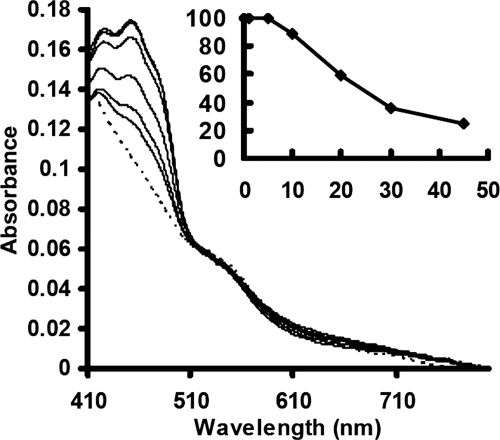
Given that ThnY does not contain a proper binding domain for these physiological reductants, as shown in the alignment of Fig. 3, we wondered whether ThnY could be reduced when NADH or NAD(P)H were used as electron donor. To do that, we have analyzed by UV-visible spectroscopy the overall process of ThnY reduction by an excess of NADH or NAD(P)H over time under anaerobic conditions (Fig. 6). The inset of Fig. 6 shows the percentage of oxidized protein over the time measured as a decrease in the absorbance observed at 450 nm and considering as 100% reduced ThnY the value obtained in the presence of sodium dithionite. These results indicate that although ThnY can apparently be reduced by pyridine nucleotides, this reduction is poor because it required at least 45 min to 1 h to achieve about 80% of the protein in its reduced form, using either NADH or NADPH. Similar results were obtained when the experiments were carried out under aerobic conditions (data not shown). This extremely slow reaction clearly indicated that neither NADH nor NADPH was effective as an electron donor for ThnY, as suspected from the poor conservation of the pyridine nucleotide binding domain shown by sequence comparison (Fig. 3C).
FIGURE 6.
Time course for the anaerobic reduction of ThnY with NAD(P)H. The spectrum of ThnY (35 μm) was recorded during 45–60 min after the addition of 20 mm of NADH. The dashed line represents ThnY fully reduced with dithionite. The insets show the percentage of oxidized ThnY, measured as the decrease in absorbance at 450 nm, over time.
Fig. 6 also indicates that reoxidation of the FAD prosthetic group was not observed under the anaerobic conditions used in the experiment. Moreover, when air was admitted to the cuvette, the spectrum of the oxidized form of the protein was not regenerated until 30 min after the oxygen was present (data not shown), thus indicating that ThnY was not reversibly autoxidized and that reduction of molecular oxygen by the enzyme was very slow. The flavin can adopt three different redox forms: the oxidized quinone form (FAD), the one-electron reduced semiquinone radical form (FADH•), and the fully reduced quinol form (FADH2) (33). The semiquinone form of the flavin was not detected at all during the course of reduction by either NADH or NADPH. This is in absolute agreement with our findings of the EPR measurement, where we could not detect any signal in the NADPH-treated sample (data not shown) but only found a clear signal resulting from the 2Fe-2S cluster in the protein sample fully reduced by dithionite (Fig. 5C). Thus, the one-electron reduced form of FAD is not stabilized in the enzyme.
Role of ThnY in Transcription Activation
The regulatory system of tetralin biodegradation genes does not work in E. coli in vivo, and we have been unable to reproduce an in vitro transcription system from the thn promoters using the E. coli RNA polymerase. Nevertheless, the initial step in transcription activation (i.e. binding of the transcriptional activator ThnR to a probe containing the thnB-thnC divergent promoters) can be studied in vitro by EMSA and DNase I footprinting (12).
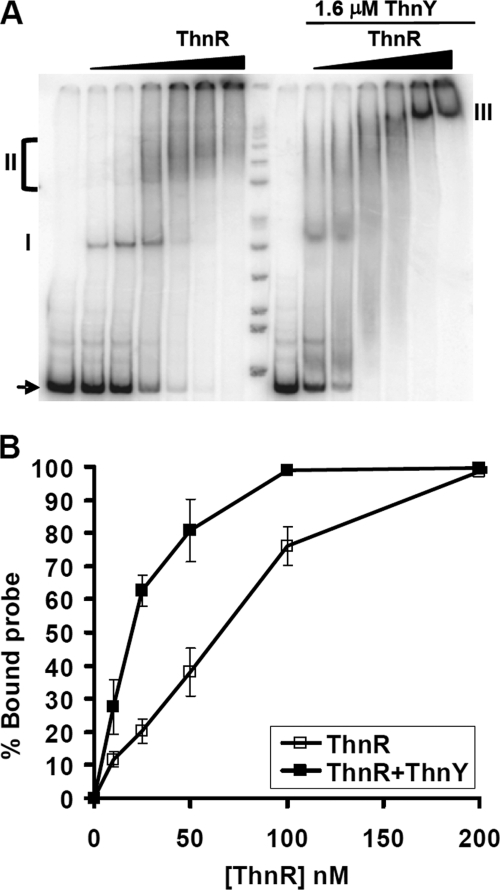
In order to detect any change in binding affinity or mobility of the protein-DNA complexes, purified ThnY-His6 was added to binding assays using a DNA probe comprising the thnB-thnC divergent promoter region. As shown previously (12), ThnR binds to two sites in this probe with different affinity. When increasing ThnR concentrations, ThnR initially binds to site C, forming the defined complex I, and subsequently binds to site B at higher concentrations to form diffused higher order complexes, labeled as II in Fig. 7A. In the presence of ThnY, unbound DNA disappears at lower ThnR concentrations. Quantification of the binding assays indicated that binding affinity increased 3.4-fold (Fig. 7B). Additionally, the diffused complexes II are substituted by a defined higher order complex III that migrates more slowly.
FIGURE 7.
Binding of ThnR to thnB-thnC promoters region. A, electrophoretic mobility shift assays of ThnR binding to the wild type thnB-thnC promoter region in the absence (left) or presence of 1.6 μm ThnY (right). The black triangles represent increasing ThnR-His6 concentrations: 0, 50, 100, 200, 400, 800, and 1200 nm ThnR tetramer. Free probes are indicated by an arrow, and the protein-DNA complexes are marked as I, II, and III. Binding was efficiently titrated out by an unlabeled wild type thnB-thnC promoter region probe. B, binding affinities of ThnR to the thnB-thnC promoter region in the presence or absence of ThnY, calculated as a percentage of the remaining unbound probe. Error bars, S.D.
These results clearly indicate that ThnY affects binding affinity of ThnR and also the structure of the protein-DNA complex. In order to characterize these changes, DNase I footprinting assays of ThnR were performed with or without ThnY. In the absence of ThnY, ThnR showed a complex pattern of protection indicating binding to different subsites and DNA looping of the intergenic region, as previously published (12). The addition of ThnY to the footprinting assay did not change at all the ThnR protection pattern in this region (data not shown). Thus, although ThnY may increase ThnR affinity and reduce mobility of the complex, ThnY does not alter the intimate interaction of ThnR with its binding sites. This also suggests that the ThnY function in activating transcription from the thnB and thnC promoters is exerted through protein-protein interactions with the activator ThnR.
DISCUSSION
Previous mutational analyses had shown that the product encoded by thnY was essential for tetralin biodegradation capacity in S. macrogolitabida strain TFA. Interestingly, although its sequence resembled those of ferredoxin reductases involved in the first step of the catabolic pathway, its role is not catalytic but regulatory. ThnY is essential for expressing the tetralin biodegradation capability, particularly for transcription from the four promoters that are induced in the presence of tetraline (5, 10, 12). In an effort to understand the role of ThnY in regulating tetralin biodegradation genes, we have analyzed its expression pattern and purified and analyzed a number of biochemical and functional characteristics of a His-tagged version of this protein.
Antibodies against ThnY that were obtained using ThnY-His6 as the antigen allowed us to make semiquantitative estimations of the in vivo ThnY concentration under different conditions. The results (Fig. 2) were fully concordant with the expression pattern exhibited by the thnR regulatory gene (12) and indicated that there was a low but detectable basal expression under non-inducing conditions, which increased in a ThnR-dependent manner when cells grew under inducing conditions. Given the close proximity of thnY to thnR, whose reading frames are separated by 7 bp, and their expression pattern, we conclude that thnR and thnY are cotranscribed. Thus, under non-inducing conditions, both regulatory genes are constitutively transcribed from PR in a short transcriptional unit comprising just thnRY, which would allow the production of sufficient amounts of the regulatory proteins to initiate the induction process in the presence of tetralin. Under inducing conditions, ThnR and ThnY would activate transcription from the highest affinity promoter, PC, which in turn results in autoactivation of thnRY by transcriptional read-through. The increase in the in vivo ThnR and ThnY concentrations would allow full activation of transcription of the remaining genes from the lower affinity promoters PB, PH, and PM (12).
Sequence analysis indicated that ThnY is most similar to ferredoxin reductases of class III, according to the biochemical classification proposed by Batie et al. (7). Enzymes of this class are components of the electron transfer system of ring-hydroxylating dioxygenases that catalyze the initial dioxygenation reaction of the aromatic benzene rings and play an important role in the degradation of various environmental pollutants. They all contain a plant type [2Fe-2S] cluster and FAD as prosthetic groups. Electrons are transferred from the pyridine nucleotides, through the FAD and the [2Fe-2S] cluster, to an intermediary Rieske type ferredoxin, which in turn transfers the electron to the dioxygenase. Purified ThnY is an orange-brown protein and contains flavin and a plant type iron-sulfur cluster as redox centers, as indicated by its distinctive visible absorbance (Fig. 4B) and EPR spectrum (Fig. 5C). The flavin, which is non-covalently bound, was identified as flavin adenine dinucleotide by using thin layer chromatography (Fig. 5B) and estimated to be in equimolecular concentration with the protein (Fig. 5A).
Both cofactors seem to be functional in ThnY because both can be readily reduced in the presence of an excess of dithionite (Figs. 4B and 5C). Of course, the observed reduction is artificial because the dithionite is fully reducing all of the reducible centers at the same time. However, because ThnY can be reduced, it is tempting to speculate that its redox state may influence its regulatory function. Regulation of gene expression mediated by transcriptional factors or modulatory proteins that function as redox-sensitive switches has been reported in different systems (34). These include essential processes, such as photosynthesis in purple and non-purple phototrophic bacteria (35, 36), response to oxidative stress (37), nitrogen fixation in free living organisms through NifL (38, 39), or anaerobic metabolism through FNR (40). The common feature in these regulators is that they control gene expression in response to the presence of oxygen or oxidative stress, and the function of some of them, like FNR, is directly regulated by oxygen. However, in the case of ThnY, oxygen (or its absence) cannot be a triggering signal because thn genes are similarly regulated in aerobiosis or in anaerobiosis.3
Unlike the typical ferredoxin reductases, which show activity in the presence of either NADH or NADPH and which are able to deliver electrons from these reductants to artificial electron acceptors, such as cytochrome c, DCPIP, or ferricyanide, purified ThnY was unable to reduce cytochrome c or ferricyanide and showed barely detectable diaphorase activity using DCPIP as the electron acceptor. The lack of these activities may be explained by the very inefficient reduction of ThnY when using any of the pyridine nucleotides as the electron donor (Fig. 6). As a ferredoxin reductase of the dioxygenase systems, ThnY should receive electrons from pyridine nucleotides. However, the ThnY amino acid sequence showed that the fingerprint sequence GGXGXXP in the C-terminal domain, which is proposed to be involved in the binding of the adenosine-5′-phosphate groups of NADH or 2′-phospho-AMP of NADPH, is not conserved in ThnY (22). In addition, where structures are available, the conserved Pro from the former signature and the Cys residue from the conserved motif (yXCGp) are shown to contact the bound nicotinamide moiety. A mutant form of a ferredoxin-NADP+ reductase with the Cys changed to Ser displayed decreased rates of flavin reduction by NADPH and lower yields of intermediate pyridine nuc1eotide:flavin charge transfer species (41). In ThnY, Phe substitutes Cys. These substitutions in the conserved NAD(P)H binding domain of ThnY may have an effect on the precise mode in which the pyridine nucleotide is bound, thus preventing ThnY from efficiently accepting electrons from it. These substitutions may actually be important for the correct function of ThnY to prevent it from being readily reduced under non-appropriate conditions if the NAD(P)H pool is sufficiently high. Thus, if the function of ThnY depends on its redox state, signals affecting thn expression can be transduced to ThnY by accumulating a so far unknown physiological ThnY reductant.
Inclusion of ThnY in the EMSAs (Fig. 7) had weak but very important effects on ThnR binding to the thnB-thnC intergenic region. In first place, protein-DNA complexes were formed with significantly higher affinity in the presence of ThnY. Second, the diffused higher order complexes observed in the absence of ThnY, which are indicative of unstable complexes (12), were substituted by a more defined band in its presence, which suggests that ThnY stabilizes the ThnR-promoter complex at this region. These effects clearly indicated a direct positive role of oxidized ThnY in regulating thn gene transcription. Given the characteristics of ThnY, its presumed function is to modulate the function of the LysR type transcriptional activator ThnR. Because the oxidized form of ThnY is active (Fig. 7), its maximum effect is observed when the ratio of ThnY monomer to ThnR tetramer is at least 2 (data not shown), and it does not change the footprint produced by ThnR by itself, we propose that its regulatory function is not catalytic but involves formation of a transcriptionally competent ThnR-ThnY complex through protein-protein interactions. Although most LysR type regulators directly sense the environmental signals to which they respond, a number of examples have been reported in which auxiliary proteins are also required to regulate transcription (18, 42–45); such a two-component composition allows the regulatory system to respond to additional signals. In the case of the thn genes, transcription is subjected to carbon catabolite repression and is also responding to a modulation system exerted by the reduced form of the ferredoxin ThnA3, which prevents thn gene induction if the inducer molecule cannot be efficiently metabolized by the tetralin biodegradation pathway, thus preventing gratuitous induction (11). These additional signals may be integrated in the regulatory system by affecting the functional state of ThnY, presumably by modifying its redox state.
In the last few years, a number of metabolic enzymes have been reported to have a direct role in regulation of both prokaryotic and eukaryotic gene expression in addition to its enzymatic activity. This principle of a dual function of one protein is envisioned as a way of linking cellular metabolism to gene expression (46–48). ThnY represents the first example of an electron transport component that has evolved to lose its capacity of accepting electrons from pyridine nucleotides and instead has been recruited to play a direct role in regulating transcription from target promoters.
Acknowledgment
We thank Dr. Reinhardt Kappl and K. M. Ewen for help with the EPR measurements.
This work was supported by Spanish Ministry of Science and Innovation Grants BIO2008-01805 and CSD2007-00005 and Andalusian Government Grants P05-CVI-131 and P07-CVI-2518 (to L.L.G.).
F. Reyes-Ramírez and E. Santero, unpublished data.
- βHB
- β-hydroxybutyrate
- DCPIP
- 2,6-dichlorophenolindophenol
- FNR
- ferredoxin:NAD(P)+ reductase.
REFERENCES
- 1. Hernáez M. J., Andújar E., Ríos J. L., Kaschabek S. R., Reineke W., Santero E. (2000) J. Bacteriol. 182, 5448–5453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andújar E., Hernáez M. J., Kaschabek S. R., Reineke W., Santero E. (2000) J. Bacteriol. 182, 789–795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Andújar E., Santero E. (2003) Microbiology 149, 1559–1567 [DOI] [PubMed] [Google Scholar]
- 4. Moreno-Ruiz E., Hernáez M. J., Martínez-Pérez O., Santero E. (2003) J. Bacteriol. 185, 2026–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. López-Sánchez A., Floriano B., Andújar E., Hernáez M. J., Santero E. (2010) Appl. Environ. Microbiol. 76, 110–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mason J. R., Cammack R. (1992) Annu. Rev. Microbiol. 46, 277–305 [DOI] [PubMed] [Google Scholar]
- 7. Batie C. J., Ballou D. P., Correll C. C. (1991) in Chemistry and Biochemistry of Flavoenzymes (Müller F. ed) pp. 543–556, CRC Press, Inc., Boca Raton, FL [Google Scholar]
- 8. Butler C. S., Mason J. R. (1997) Adv. Microb. Physiol. 38, 47–84 [DOI] [PubMed] [Google Scholar]
- 9. Ferraro D. J., Gakhar L., Ramaswamy S. (2005) Biochem. Biophys. Res. Commun. 338, 175–190 [DOI] [PubMed] [Google Scholar]
- 10. Martínez-Pérez O., Moreno-Ruiz E., Floriano B., Santero E. (2004) J. Bacteriol. 186, 6101–6109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Martínez-Pérez O., López-Sánchez A., Reyes-Ramírez F., Floriano B., Santero E. (2007) J. Bacteriol. 189, 3768–3775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. López-Sánchez A., Rivas-Marín E., Martínez-Pérez O., Floriano B., Santero E. (2009) Mol. Microbiol. 73, 1086–1100 [DOI] [PubMed] [Google Scholar]
- 13. Sambrook J., Russell D.W. (2001) Molecular Cloning: A Laboratory Manual, 3rd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 14. Dorn E., Hellwig M., Reineke W., Knackmuss H. J. (1974) Arch. Microbiol. 99, 61–70 [DOI] [PubMed] [Google Scholar]
- 15. Hernáez M. J., Reineke W., Santero E. (1999) Appl. Environ. Microbiol. 65, 1806–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Miller J. H. (1992) A Short Course in Bacteriol genetics: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 17. Frago S., Martínez-Júlvez M., Serrano A., Medina M. (2008) BMC Microbiol. 8, 160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Porrúa O., García-Jaramillo M., Santero E., Govantes F. (2007) Mol. Microbiol. 66, 410–427 [DOI] [PubMed] [Google Scholar]
- 19. Lane D., Cavaillé J., Chandler M. (1994) J. Mol. Biol. 242, 339–350 [DOI] [PubMed] [Google Scholar]
- 20. Karlsson A., Beharry Z. M., Matthew Eby D., Coulter E. D., Neidle E. L., Kurtz D. M., Jr., Eklund H., Ramaswamy S. (2002) J. Mol. Biol. 318, 261–272 [DOI] [PubMed] [Google Scholar]
- 21. Kauppi B., Lee K., Carredano E., Parales R. E., Gibson D. T., Eklund H., Ramaswamy S. (1998) Structure 6, 571–586 [DOI] [PubMed] [Google Scholar]
- 22. Karplus P. A., Daniels M. J., Herriott J. R. (1991) Science 251, 60–66 [PubMed] [Google Scholar]
- 23. Aliverti A., Pandini V., Pennati A., de Rosa M., Zanetti G. (2008) Arch. Biochem. Biophys. 474, 283–291 [DOI] [PubMed] [Google Scholar]
- 24. Ceccarelli E. A., Arakaki A. K., Cortez N., Carrillo N. (2004) Biochim. Biophys. Acta 1698, 155–165 [DOI] [PubMed] [Google Scholar]
- 25. Correll C. C., Ludwig M. L., Bruns C. M., Karplus P. A. (1993) Protein Sci. 2, 2112–2133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Macherrex P. (1999) in Flavoprotein Protocols (Chapman S. K., Reid G. A.) (eds), pp. 1–7, Humana Press, Totowa, NJ [Google Scholar]
- 27. Ewen K. M., Kleser M., Bernhardt R. (2011) Biochim. Biophys. Acta 1814, 111–125 [DOI] [PubMed] [Google Scholar]
- 28. Yamaguchi M., Fujisawa H. (1980) J. Biol. Chem. 255, 5058–5063 [PubMed] [Google Scholar]
- 29. Haigler B. E., Gibson D. T. (1990) J. Bacteriol. 172, 457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fetzner S., Müller R., Lingens F. (1992) J. Bacteriol. 174, 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Nam J. W., Nojiri H., Noguchi H., Uchimura H., Yoshida T., Habe H., Yamane H., Omori T. (2002) Appl. Environ. Microbiol. 68, 5882–5890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lovenberg W. (1973) Iron-Sulfur Proteins: Molecular Properties, pp. 214–222, Academic Press, Inc., New York [Google Scholar]
- 33. Massey V. (2000) Biochem. Soc. Trans. 28, 283–296 [PubMed] [Google Scholar]
- 34. Bauer C. E., Elsen S., Bird T. H. (1999) Annu. Rev. Microbiol. 53, 495–523 [DOI] [PubMed] [Google Scholar]
- 35. Elsen S., Jaubert M., Pignol D., Giraud E. (2005) Mol. Microbiol. 57, 17–26 [DOI] [PubMed] [Google Scholar]
- 36. Dubbs J. M., Tabita F. R. (2004) FEMS Microbiol. Rev. 28, 353–376 [DOI] [PubMed] [Google Scholar]
- 37. Zheng M., Storz G. (2000) Biochem. Pharmacol. 59, 1–6 [DOI] [PubMed] [Google Scholar]
- 38. Hill S., Austin S., Eydmann T., Jones T., Dixon R. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 2143–2148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Little R., Reyes-Ramirez F., Zhang Y., van Heeswijk W. C., Dixon R. (2000) EMBO J. 19, 6041–6050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Unden G., Schirawski J. (1997) Mol. Microbiol. 25, 205–210 [DOI] [PubMed] [Google Scholar]
- 41. Aliverti A., Piubelli L., Zanetti G., Lübberstedt T., Herrmann R. G., Curti B. (1993) Biochemistry 32, 6374–6380 [DOI] [PubMed] [Google Scholar]
- 42. Heil G., Stauffer L. T., Stauffer G. V. (2002) Microbiology 148, 2203–2214 [DOI] [PubMed] [Google Scholar]
- 43. Huang J., Yindeeyoungyeon W., Garg R. P., Denny T. P., Schell M. A. (1998) J. Bacteriol. 180, 2736–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kovacikova G., Lin W., Skorupski K. (2004) Mol. Microbiol. 53, 129–142 [DOI] [PubMed] [Google Scholar]
- 45. García-González V., Jiménez-Fernández A., Hervás A. B., Canosa I., Santero E., Govantes F. (2009) FEMS Microbiol. Lett. 300, 222–229 [DOI] [PubMed] [Google Scholar]
- 46. Hall D. A., Zhu H., Zhu X., Royce T., Gerstein M., Snyder M. (2004) Science 306, 482–484 [DOI] [PubMed] [Google Scholar]
- 47. Shi Y., Shi Y. (2004) Trends Genet. 20, 445–452 [DOI] [PubMed] [Google Scholar]
- 48. Zhou Y., Zhu W., Bellur P. S., Rewinkel D., Becker D. F. (2008) Amino Acids 35, 711–718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Hanahan D. (1983) J. Mol. Biol. 166, 557–580 [DOI] [PubMed] [Google Scholar]
- 50. Govantes F., Molina-López J. A., Santero E. (1996) J. Bacteriol. 178, 6817–6823 [DOI] [PMC free article] [PubMed] [Google Scholar]