Abstract
The Bacillus subtilis redox regulator Fnr controls genes of the anaerobic metabolism in response to low oxygen tension. An unusual structure for the oxygen-sensing [4Fe-4S]2+ cluster was detected by a combination of genetic experiments with UV-visible and Mössbauer spectroscopy. Asp-141 was identified as the fourth iron-sulfur cluster ligand besides three Cys residues. Exchange of Asp-141 with Ala abolished functional in vivo complementation of an fnr knock-out strain by the mutagenized fnr gene and in vitro DNA binding of the recombinant regulator FnrD141A. In contrast, substitution of Asp-141 with Cys preserved [4Fe-4S]2+ structure and regulator function.
Keywords: Bacterial Signal Transduction, DNA-binding Protein, Iron-Sulfur Protein, Mossbauer Spectroscopy, UV Spectroscopy, Bacillus subtilis, Fnr, Anaerobiosis, Redox Regulator
Introduction
Eukaryotic life is adapted to an oxygen-dependent energy generation. In contrast, many prokaryotes populate their ecological niches by the ability to survive and multiply under conditions of low oxygen tension. Anaerobic growth of these organisms is mediated by a broad spectrum of proton gradient-generating alternative respiration strategies. In addition, substrate level phosphorylation employing highly diverse fermentation processes allows for ATP formation and growth under anaerobic conditions. Moreover, this anaerobic energy metabolism was found to be essential for many pathogenic microorganisms during infection and biofilm formation. Because of the better energy yield, organisms prefer oxygen-dependent growth over anaerobic energy generation strategies. Additionally, several components of the anaerobic respiratory machineries were found to be oxygen-sensitive. Consequently, a strict regulation of the aerobic-anaerobic transition is advised. For oxygen sensing, many different types of sensors have been identified in bacteria and archaea, most of which react directly with oxygen by oxygen-reactive groups, like heme, FeS clusters, cysteine pairs, and FAD (1).
A key regulatory protein in bacteria is Fnr, named after the fumarate and nitrate reduction-negative phenotype of an fnr gene-defective Escherichia coli strain (2). The N-terminal part of the E. coli Fnr protein contains four cysteine residues, Cys-20, Cys-23, Cys-29, and Cys-122, which are essential for Fnr function by coordinating a [4Fe-4S]2+ cluster responsible for oxygen sensing. The C-terminal DNA-binding domain recognizes specific binding sites located in Fnr-controlled promoters (2).
In contrast to E. coli Fnr, the [4Fe-4S]2+ cluster of the redox regulator from the Gram-positive model bacterium Bacillus subtilis possesses only three cysteine ligands (Cys-227, Cys-230, and Cys-235) localized at the C terminus of the protein and a fourth unknown non-cysteinyl ligand (3). In most cases, the iron atoms of [4Fe-4S]2+ clusters are coordinated by four cysteine residues as found for E. coli Fnr. However, there are reported precedents of iron-sulfur clusters with only three cysteine ligands. Histidine residues are known to be involved in iron-sulfur cluster coordination in so-called “Rieske clusters” that are usually part of membrane-localized electron-transferring proteins (4). However, alanine substitutions of all conserved histidine residues of B. subtilis Fnr failed to abolish Fnr function and excluded histidines as the fourth ligand (3).
EXPERIMENTAL PROCEDURES
Bacterial Strains, Growth Conditions, and β-Galactosidase Assays
Anaerobic growth of various B. subtilis strains and β-galactosidase assays were performed as described previously (3).
Generation of B. subtilis Mutant Fnr Proteins and Their Production, Purification, and Biochemical Characterization
The integrative vector pHRB1 was used for the chromosomal integration of mutant fnr genes at the amyE locus of B. subtilis JH642. There, the fnr genes were expressed under the control of the xylose-inducible xylA promoter (3). The fnr mutants were obtained by site-directed mutagenesis using the QuikChange kit (Stratagene, Heidelberg, Germany). Primers used for mutagenesis are listed in supplemental Table S1. Construction of the expression vectors for the mutant fnr genes and the expression and affinity purification of the mutant Fnr proteins were performed as described previously (3). Protein concentrations were determined using the 2D-Quant kit (Amersham Biosciences). BSA served as a standard. Iron and sulfide determinations were performed as described (5, 6). Iron standards were obtained from Merck (Darmstadt, Germany). UV-visible light spectra of purified Fnr proteins were recorded using a Lambda 2 spectrophotometer (PerkinElmer Instruments, Überlingen, Germany). Measurements were performed under strict anaerobic conditions.
For DNase I footprinting, a 310-bp narG promoter fragment corresponding to positions −210 to +100 with respect to the transcription start site of narG was amplified by PCR using a digoxigenin-labeled primer and an unlabeled primer. The following primer sets were used: 1) digoxigenin-EH176 and EH177 and 2) EH176 and digoxigenin-EH177 (supplemental Table S1). The footprint analysis was performed as described previously (7).
Preparation of E. coli Cell-free Extracts Containing B. subtilis Fnr Proteins for Mössbauer Spectroscopy
Strains containing plasmid-encoded fnr, fnrD141A, or fnrD141C hosted by the vector pASK-IBA45-plus (IBA, Göttingen, Germany) in E. coli BL21-CodonPlus(DE3)-RIL cells (Stratagene, La Jolla, CA) were grown in iron-free Spizizen's minimal medium supplemented with 34 μm 57FeCl3. Recombinant protein production was performed as described previously (3). After protein production for 16 h, cells were incubated in an anaerobic chamber for 3 h. Cells were harvested, and the resulting bacterial cell pellet was washed with ice-cold anaerobic buffer (100 mm Tris-HCl, 150 mm NaCl (pH 8.0), 10% glycerol, and 5 mm DTT) and centrifuged at 25,000 × g for 30 min to form a dense cell pellet. Approximately 800 mg of cell paste was transferred to a Mössbauer cup and frozen.
Mössbauer Spectroscopy
Mössbauer spectra were recorded using a spectrometer in the constant acceleration mode. Isomer shifts are given relative to α-iron at room temperature. The spectra were obtained in an Oxford continues flow cryostat. The spectra were analyzed by least-square fits using Lorentzian line shapes.
RESULTS
Asp-141 Is the Fourth Ligand of the [4Fe-4S]2+ Cluster of B. subtilis Fnr
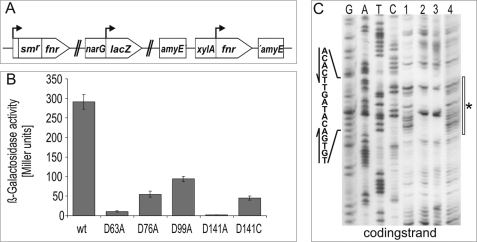
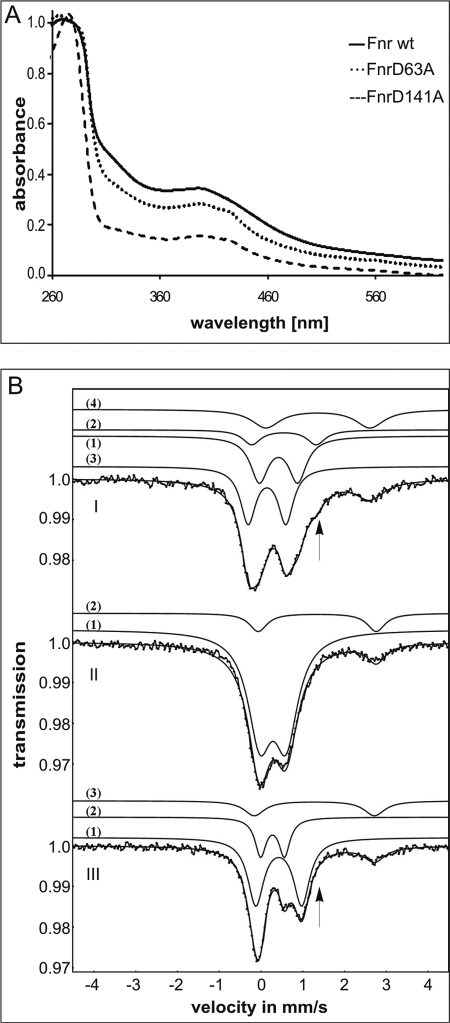
The nature of the fourth ligand of the [4Fe-4S]2+ cluster of B. subtilis Fnr, besides the three cysteine residues localized at the C terminus, was unknown. The involvement of histidine residues was excluded (3). In the [4Fe-4S]2+ cluster of a Pyrococcus furiosus ferredoxin, one of the iron atoms has an aspartate ligand in place of a cysteine (8). Recently, an aspartate ligand was also found in the protochlorophyllide oxidoreductases (DPOR) of Rhodobacter capsulatus and Thermosynechococcus (9, 10). B. subtilis Fnr possesses 15 aspartate residues. Four of them, Asp-63, Asp-76, Asp-99, and Asp-141, are highly conserved among Fnr proteins of different bacilli (3). Consequently, these four residues were individually mutated to alanine residues, and the functions of the corresponding mutant Fnr proteins were tested for their ability to complement a B. subtilis mutant fnr strain in vivo. For this purpose, the fnr-dependent anaerobic activation of the narG promoter was tested. The narGHJI operon encodes the alternative respiratory enzyme nitrate reductase. Fnr function was monitored by measuring β-galactosidase activities derived from a narG-lacZ reporter gene fusion under nitrate respiratory conditions (Fig. 1A) (3). Only exchange of Asp-63 and Asp-141 for alanine, respectively, resulted in a drastic reduction in β-galactosidase activities to values below 5% of the wild-type level (Fig. 1B). To test whether Asp-63 or Asp-141 is involved in formation of the [4Fe-4S]2+ cluster, the mutant proteins FnrD64A and FnrD141A were produced recombinantly and purified anaerobically to apparent homogeneity, and UV-visible spectra were recorded under anoxic conditions. B. subtilis wild-type Fnr protein showed an absorption shoulder at 320 nm and a broad peak with an absorption maximum around 420 nm, typical for [4Fe-4S]2+ cluster-containing proteins (Fig. 2A). In addition, an iron content of 3.6 mol of iron/mol of Fnr was determined. Similar values were obtained for FnrD63A. In contrast, FnrD141A showed only weak absorption at 420 nm and a low iron content of 1.8 mol of iron/mol of Fnr (Fig. 2A). From these results, we concluded that FnrD141A is no longer able to coordinate an intact [4Fe-4S]2+ cluster.
FIGURE 1.
Functional analyses of B. subtilis Fnr and mutant derivatives for in vivo complementation of a mutant fnr strain and in vitro promoter binding. A, shown are relevant genomic structures of B. subtilis strain HRB3 used for in vivo complementation experiments. The chromosomal fnr copy is inactivated by insertion of a spectinomycin resistance cassette. The Fnr-dependent narG promoter is fused to the reporter gene lacZ, and the tested fnr gene integrated into the amyE locus is expressed under the control of the xylA promoter. B, Fnr function was monitored by expression of an Fnr-dependent narG-lacZ reporter gene fusion under nitrate respiratory growth conditions. Corresponding β-galactosidase activities are given in Miller units. C, a DNase I footprint experiment using a narG promoter fragment representing the coding strand was performed without the addition of protein (lane 1) or with the addition of 1.6 μm FnrCCC (lane 2), FnrD63A (lane 3), and FnrD141A (lane 4). DNA sequencing ladders were included to localize the Fnr-binding sites. The white bar indicates the protected region, and the DNase I-hypersensitive site is marked by an asterisk.
FIGURE 2.
Spectroscopic analysis of B. subtilis Fnr variants. A, UV-visible spectra of anoxically purified B. subtilis wild-type Fnr (solid line), FnrD63A (dotted line), and FnrD141A (dashed line). B, Mössbauer spectra recorded at 77 k of B. subtilis wild-type Fnr (panel I), FnrD141A (panel II), and FnrD141C (panel III) in whole cell extracts of producing E. coli cells. The results of least square fit analyses represented by the solid traces of the subspectra are described in the text, and parameters are given in Table 1.
DNA binding of B. subtilis Fnr requires the presence of an intact [4Fe-4S]2+ cluster (3). Consequently, DNase I footprint analyses with a narG promoter fragment carrying a high-affinity Fnr-binding site (TGTGA–TA–TCACA) using purified FnrD63A, FnrD141A, and FnrCCC proteins were carried out. FnrCCC was used as the wild-type control and is a variant carrying solely the three cysteines (Cys-227, Cys-230, and Cys-235) required for [4Fe-4S]2+ cluster formation. Other cysteine residues not involved in [4Fe-4S]2+ cluster formation were changed to alanine residues. Although showing identical functional features to the wild-type protein, this variant revealed increased in vitro stability (3). A specific footprint indicated by a DNase I-hypersensitive site flanked by two protected regions was observed for FnrCCC and FnrD63A (Fig. 1C). In clear contrast, no protection of the DNA from DNase I digestion was visible after incubation of the promoter fragment with the FnrD141A protein, indicating that the FnrD141A protein was not able to bind to the DNA (Fig. 1C). The results of these functional DNA binding assays in combination with the iron-sulfur cluster analyses point toward a role of Asp-141 as the fourth ligand of the [4Fe-4S]2+ cluster in B. subtilis Fnr.
Replacement of Asp-141 with Cysteine Enables [4Fe-4S]2+ Cluster Formation
To finally prove Asp-141 function in [4Fe-4S]2+ cluster coordination, the FnrD141C protein was generated. The rationale of this approach was to restore [4Fe-4S]2+ cluster formation via the introduction of an alternative functional fourth ligand, in this case, a cysteine residue. In the in vivo complementation assay, FnrD141C revealed significant Fnr activity (Fig. 1B). Moreover, clear differences in the Mössbauer spectra of B. subtilis wild-type Fnr protein, FnrD141A, and FnrD141C were expected. An aspartate instead of a cysteine ligand for a [4Fe-4S]2+ cluster clearly should change the Mössbauer spectrum. To prevent the partial loss of labile FeS clusters of FnrD141C during protein purification, we measured the Mössbauer spectra of E. coli whole cell extracts carrying recombinant Fnr proteins at 77 K. The 57Fe-based Mössbauer spectroscopic approach using E. coli whole cell extracts was employed previously to study the in vivo behavior of overproduced E. coli Fnr protein (11). The Mössbauer spectra of B. subtilis wild-type Fnr revealed a subset of two quadrupole doublets with an area ratio of 3:1 for cysteine-ligated and non-cysteine-ligated iron sites characteristic for a [4Fe-4S]2+ cluster, representing 32% of the overall iron content in the probe (Fig. 2B, panel I, traces 1 and 2; and Table 1). The motif of the two quadrupole doublets formed an extra shoulder at 1.3 mm/s (Fig. 2B, panel I, arrow). The isomer shift of the non-cysteine ligand component of B. subtilis Fnr was ∼0.1 mm/s higher than that of the cysteine ligand component, indicating that the corresponding iron is coordinated to a less covalently bound ligand than sulfur, i.e. oxygen, as it is in Asp-141 (12). This result corresponds to a previously recorded Mössbauer spectrum of anaerobically purified FnrCCC protein (3). Another subspectrum representing 45% of the iron was characteristic for ferric high-spin tetrahedrally sulfur-coordinated iron sites as observed in [2Fe-2S]2+ clusters (Fig. 2B, panel I, trace 3; and Table 1). The other parameters found were similar to those observed previously for the Fe2+ metabolites that appeared after feeding E. coli cells with iron citrate (Fig. 2B, panel I, trace 4; and Table 1) (13). The occurrence of a recorded [2Fe-2S]2+ spectrum was the first indication that inactivation of [4Fe-4S]2+ of B. subtilis Fnr by oxygen may proceed via conversion to [2Fe-2S]2+. For E. coli Fnr, it was described that the functional [4Fe-4S]2+ clusters are converted to inactive [2Fe-2S]2+ clusters via oxygen contact. This triggers conformational changes that induce monomerization of the Fnr molecules, preventing DNA binding and interactions with RNA polymerase (11, 14, 15).
TABLE 1.
Quadrupole splittings and isomer shifts of Fnr samples
Spectra were analyzed by least-square fits of Lorentzian line shapes to experimental data. Panels and traces refer to Fig. 2.
| Subspectra | Component | δa | ΔEQ | Relative contribution |
|---|---|---|---|---|
| mm/s | mm/s | % | ||
| Panel I | ||||
| WT Fnr trace 1 | [4Fe-4S]2+ site 1 | 0.42 | 0.92 | 33 |
| WT Fnr trace 2 | [4Fe-4S]2+ site 2 | 0.55 | 1.54 | 11 |
| WT Fnr trace 3 | Ferric high-spin, Fe3+ | 0.14 | 0.90 | 35 |
| WT Fnr trace 4 | Ferrous high-spin, Fe2+ | 1.37 | 2.49 | 21 |
| Panel II | ||||
| FnrD141A trace 1 | Ferric high-spin, Fe3+ | 0.29 | 0.63 | 88 |
| FnrD141A trace 2 | Ferrous high-spin, Fe2+ | 1.35 | 2.83 | 12 |
| Panel III | ||||
| FnrD141C trace 1 | [4Fe-4S]2+ | 0.43 | 1.10 | 60 |
| FnrD141C trace 2 | Ferric high-spin, Fe3+ | 0.28 | 0.57 | 23 |
| FnrD141C trace 3 | Ferrous high-spin, Fe2+ | 1.28 | 2.88 | 17 |
a Isomer shifts are given relative to α-iron at room temperature.
The Mössbauer spectrum of FnrD141A did not show any signature for the presence of a [4Fe-4S]2+ cluster. The parameters for the major amount of iron in this sample were characteristic for ferric high-spin iron sites of the Fe3+ type. The rest of the iron in this sample exhibited parameters typical for ferrous high-spin iron of the Fe2+ type (Fig. 2B, panel II, traces 1 and 2; and Table 1). Thus, the D141A mutation within B. subtilis Fnr abolished coordination of the [4Fe-4S]2+ cluster and thereby inactivated the transcriptional regulator.
In contrast, the Mössbauer spectrum of FnrD141C exhibited a single set of parameters typical for four cysteine-coordinated [4Fe-4S]2+ clusters (Fig. 2B, panel III, trace 1; and Table 1). Again, subspectra typical for Fe3+ and Fe2+ ferrous high-spin iron sites were recorded to a minor extent, representing iron associated with cellular proteins (Fig. 2B, panel III, traces 2 and 3; and Table 1). Mutational analysis and in vivo activity measurements in combination with UV-visible and Mössbauer spectroscopic analyses unambiguously identified Asp-141 of B. subtilis Fnr as the fourth ligand of the [4Fe-4S]2+ cluster.
DISCUSSION
Anaerobic activation of Fnr is mediated by [4Fe-4S]2+ cluster formation between Cys-227, Cys-230, Cys-235, and Asp-141. This is one of the rare examples where aspartate serves as a ligand for [4Fe-4S]2+ coordination. The Thermococcus profundus ferredoxin was one of the first described [4Fe-4S] clusters ligated by an aspartate (16). The cluster was described as highly sensitive to oxygen. A solution NMR-based molecular model of the related [4Fe-4S] cluster of a P. furiosus ferredoxin led to the assumption that an aspartate ligation places stress on the cluster and renders it more reducible (17). A comparative computational investigation of the reduction potentials of partly aspartate- and fully cysteine-ligated [4Fe-4S] clusters concluded that the aspartate ligation facilitates the reversibility of the redox reaction and renders reduction and oxidation easier (18). Interestingly, the [4Fe-4S]2− cluster assembly protein IscA from Acidithiobacillus ferrooxidans also employs an aspartate ligand for its [4Fe-4S]2− cluster, obviously to allow enhanced transfer and assembly (19). Finally, the highly oxygen-sensitive chlorophyll biosynthetic dark-operative enzyme protochlorophyllide oxidoreductase utilizes an aspartate-ligated intersubunit cluster for enzyme assembly and redox chemistry (9, 10).
One can speculate that the aspartate-ligated [4Fe-4S]2− cluster provides the dimeric B. subtilis Fnr protein with a highly redox-sensitive, fast reactive, and rearrangement-friendly sensory unit. In contrast, the E. coli counterpart, carrying a fully cysteine-ligated cluster, follows a different regulatory strategy. Here, the monomeric, inactive regulator gets dimerized and activated upon anaerobic formation of intact [4Fe-4S]2+ cluster.
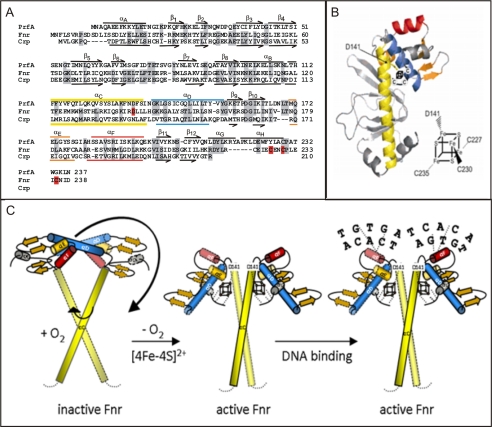
Fnr of B. subtilis is homologous to the structurally well characterized cAMP receptor protein Crp of E. coli. In addition, Listeria monocytogenes PrfA shares 23% sequence identity with B. subtilis Fnr. A model of active B. subtilis Fnr using SWISS-MODEL, a server for automated comparative modeling of three-dimensional protein structures (ExPASy), was created based on the PrfA-G145S structure (Fig. 3, A and B) (19). According to amino acid sequence alignments, secondary structure prediction, and three-dimensional modeling of B. subtilis Fnr, Asp-141 is located at the C-terminal end of helix C (Fig. 3). Analysis of the crystal structures of apo-Crp and cAMP-activated Crp bound to DNA revealed that large conformational changes in the protein dimer were necessary to allow DNA binding (16). It was suggested that cAMP stabilizes the active DNA-binding conformation of Crp through the interactions of N-6 of the adenosine moiety of cAMP with helices C. As a consequence, binding of cAMP thereby results in a coil-to-helix transition that extends the coiled-coil dimerization interface of helix C by three turns and concomitantly causes rotation of the DNA-binding domains of Crp. As a result of this rotation, the DNA recognition helices of the helix-turn-helix motif gets the correct orientation to interact with DNA (17). The structure of a constitutively active PrfA mutant of L. monocytogenes, PrfA-G145S, is comparable with the structure of active Crp (18). Assuming a similar activation mechanism for B. subtilis Fnr as described for Crp via the rearrangement of the DNA-binding domain, this active B. subtilis Fnr form can be induced by the coordination of the [4Fe-4S]2+ cluster with the participation of Asp-141 in helix C (Fig. 3C).
FIGURE 3.
Proposed location and function of the [4Fe-4S]2+ cluster using a structural model of B. subtilis Fnr. A, sequence and structure comparison of B. subtilis Fnr with L. monocytogenes PrfA and E. coli Crp. Identical residues are shaded in gray, and ligands of the [4Fe-4S]2+ cluster are indicated in red. The secondary structure elements of PrfA and Crp are schematically shown above and below the alignment: α-helices are represented as lines and β-strands as arrows. B, model of the B. subtilis Fnr monomer based on the structure of the constitutively active PrfA mutant PrfA-G145S (Protein Data Bank code 2BGC) using SWISS-MODEL (ExPASy). The C-terminal amino acid residues unique to Fnr are shown as an extension (black line). This region contains three cysteine residues (Cys-227, Cys-230, and Cys-235) coordinating the [4Fe-4S]2+ cluster together with Asp-141. C, model of [4Fe-4S]2+ cluster-dependent activation of Fnr. Under aerobic conditions (+ O2), dimeric Fnr does not carry an intact [4Fe-4S]2+ cluster. Anoxic conditions allow FeS cluster formation with subsequent movement of helix D. This novel conformation is the prerequisite for promoter binding. (The N-terminal part of Fnr is not represented in the model.)
Supplementary Material
Footnotes
This work was supported by Deutsche Forschungsgemeinschaft Grant Ha3456-1/3 and the Fonds der Chemischen Industrie.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1.
REFERENCES
- 1. Green J., Crack J. C., Thomson A. J., LeBrun N. E. (2009) Curr. Opin. Microbiol. 12, 145–151 [DOI] [PubMed] [Google Scholar]
- 2. Kiley P. J., Beinert H. (2003) Curr. Opin. Microbiol. 6, 181–185 [DOI] [PubMed] [Google Scholar]
- 3. Reents H., Gruner I., Harmening U., Böttger L. H., Layer G., Heathcote P., Trautwein A. X., Jahn D., Härtig E. (2006) Mol. Microbiol. 60, 1432–1445 [DOI] [PubMed] [Google Scholar]
- 4. Britt R. D., Sauer K., Klein M. P., Knaff D. B., Kriauciunas A., Yu C. A., Yu L., Malkin R. (1991) Biochemistry 30, 1892–1901 [DOI] [PubMed] [Google Scholar]
- 5. Lovenberg W., Buchanan B. B., Rabinowitz J. C. (1963) J. Biol. Chem. 238, 3899–3913 [PubMed] [Google Scholar]
- 6. Beinert H. (1983) Anal. Biochem. 131, 373–378 [DOI] [PubMed] [Google Scholar]
- 7. Heroven A. K., Nagel G., Tran H. J., Parr S., Dersch P. (2004) Mol. Microbiol. 53, 871–888 [DOI] [PubMed] [Google Scholar]
- 8. Calzolai L., Gorst C. M., Zhao Z. H., Teng Q., Adams M. W., La Mar G. N. (1995) Biochemistry 34, 11373–11384 [DOI] [PubMed] [Google Scholar]
- 9. Bröcker M. J., Schomburg S., Heinz D. W., Jahn D., Schubert W. D., Moser J. (2010) J. Biol. Chem. 285, 27336–27345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Muraki N., Nomata J., Ebata K., Mizoguchi T., Shiba T., Tamiaki H., Kurisu G., Fujita Y. (2010) Nature 465, 110–114 [DOI] [PubMed] [Google Scholar]
- 11. Popescu C. V., Bates D. M., Beinert H., Münck E., Kiley P. J. (1998) Proc. Natl. Acad. Sci. U.S.A. 95, 13431–13435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schünemann V., Winkler H. (2000) Rep. Prog. Phys. 63, 263–353 [Google Scholar]
- 13. Matzanke B. F., Müller G. I., Bill E., Trautwein A. X. (1989) Eur. J. Biochem. 183, 371–379 [DOI] [PubMed] [Google Scholar]
- 14. Crack J., Green J., Thomson A. J. (2004) J. Biol. Chem. 279, 9278–9286 [DOI] [PubMed] [Google Scholar]
- 15. Sutton V. R., Mettert E. L., Beinert H., Kiley P. J. (2004) J. Bacteriol. 186, 8018–8025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Imai T., Taguchi K., Ogawara Y., Ohmori D., Yamakura F., Ikezawa H., Urushiyama A. (2001) J. Biochem. 130, 649–655 [DOI] [PubMed] [Google Scholar]
- 17. Sharma H., Yu S., Kong J., Wang J., Steitz T. A. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 16604–16609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Popovych N., Tzeng S. R., Tonelli M., Ebright R. H., Kalodimos C. G. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6927–6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Eiting M., Hagelüken G., Schubert W. D., Heinz D. W. (2005) Mol. Microbiol. 56, 433–446 [DOI] [PubMed] [Google Scholar]
- 20. Deleted in proof.
- 21. Deleted in proof.
- 22. Deleted in proof.
- 23. Deleted in proof.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.