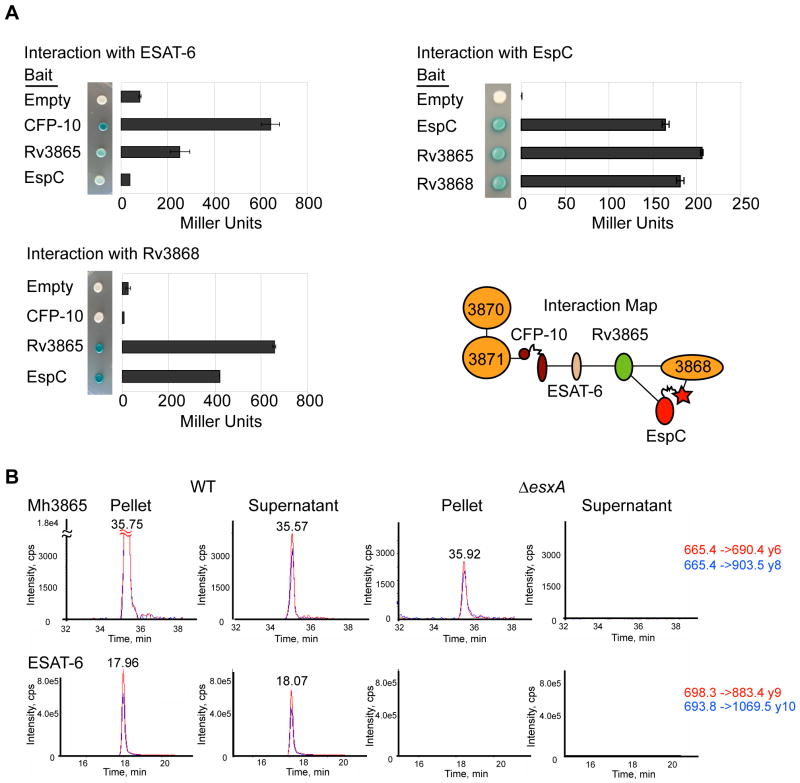
Fig. 4. At least three ESX-1 substrates interact for form a multi-protein complex in the cytosol.
(A) Yeast two-hybrid analysis measuring the interaction of M. tuberculosis ESX-1 substrates with Rv3868. Error bars represent standard deviation. Interaction map of ESX-1 associated AAA ATPases and substrates is shown. (B) Mh3865 (EspFMm) is an ESX-1 substrate. Shown are the MRM elution profiles for EspFMm and ESAT-6Mm peptides in digested M. marinum pellets (P) or supernatants (S). Two transitions (Red&Blue) for each peptide in WT or ΔesxA M. marinum cells are shown. For Mh3865 (EspFMm) and ESAT-6Mm, the transitions are shown as parent mass (m/z) -> fragment mass (m/z), and are shown to the right in red and blue. Additional peptides, including those for GroES and confirmatory spectra are in the supplementary data (Figures S6 and S7).