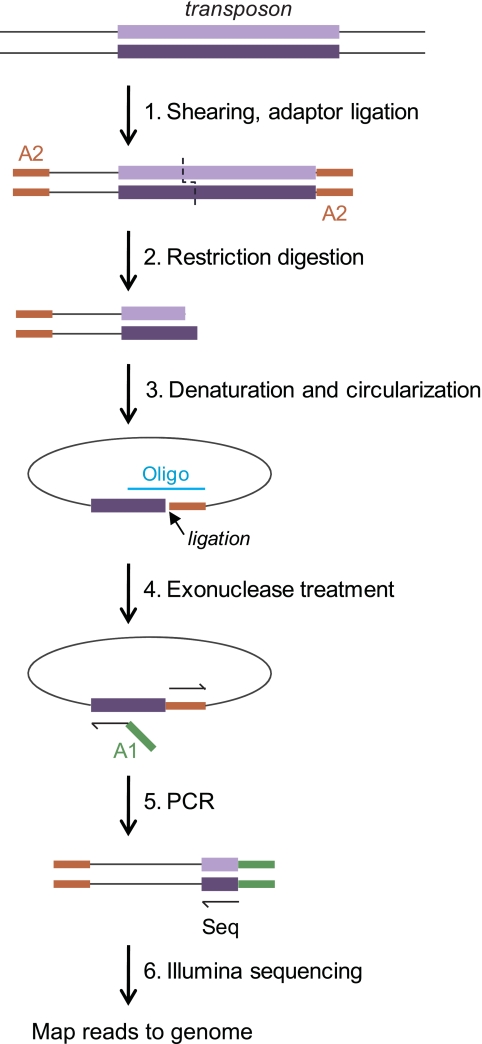
FIG 1 .
Tn-seq circle method. The steps used to amplify and sequence transposon insertion junctions are illustrated, beginning with a DNA fragment carrying a transposon insertion (top). First, total DNA from a mutant pool is sheared and end repaired, and one Illumina adaptor (A2) is ligated to all free ends (step 1). The sample is then digested with a restriction enzyme that cuts near one transposon end (in this work, BamHI, which cuts 114 bp from the transposon’s left end) (step 2). Following a size selection step, single-strand fragments which include the transposon end are circularized by templated ligation (step 3). Oligo, oligonucleotide. Fragments which have not circularized (representing most of the DNA in the sample) are degraded in a subsequent exonuclease step (step 4). The transposon-genome junctions from the circularized fragments are then amplified by quantitative PCR in a step in which the second required Illumina adaptor (A1) is introduced (step 5). The products are sequenced on an Illumina flow cell using a sequencing primer corresponding to the transposon end (Seq), and each sequence read is then mapped to the genome (step 6).