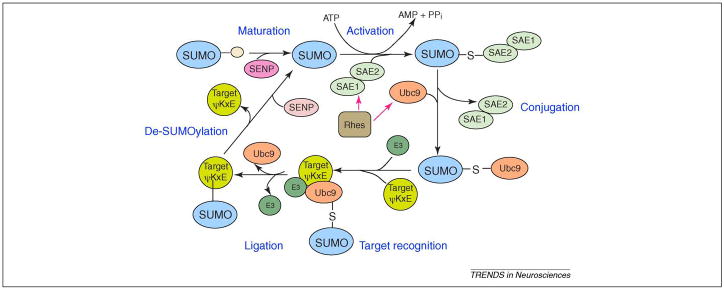
Figure 2.
The SUMO modification cycle. SUMO precursor proteins contain a C-terminal extension which is cleaved by SUMO-specific carboxyl-terminal hydrolase activity of a sentrin-specific protease (SENP) enzyme to produce a carboxyl-terminal diglycine motif in a process called ‘SUMO maturation’. The mature SUMO is activated by an E1 SUMO activating enzyme (SAE1/2) which utilizes ATP and links the C-terminal glycine of SUMO by a thioester bond to a cysteine in the E1 activating enzyme. During the conjugation step, the SUMO moiety is transferred from the E1 activating enzyme to Ubc9, the SUMO E2 enzyme, which then attaches the SUMO to a lysine residue in the target protein that is typically, but not always, found within the consensus sequence ΨKxE (Ψ represents hydrophobic amino acids). SUMO E3 proteins stimulate protein sumoylation by associating with both UBC9 and substrates to increase the efficiency of the modification reaction by a process called target recognition and ligation. The SUMO E3 proteins identified to date include members of the family of Protein Inhibitor of Activated STAT (PIAS) family of proteins (PIAS1, PIAS3, PIASx and PIASy), the polycomb protein 2 (Pc2), and Ran-binding protein 2 (RANBP2). SUMO groups are removed from proteins by SENPs, of which there are six human forms. Rhes, a small G-protein, is selectively localized to the corpus striatum and binds directly to both E1 and Ubc9, enhancing cross-sumoylation as well as thioester transfer from E1 to Ubc9.