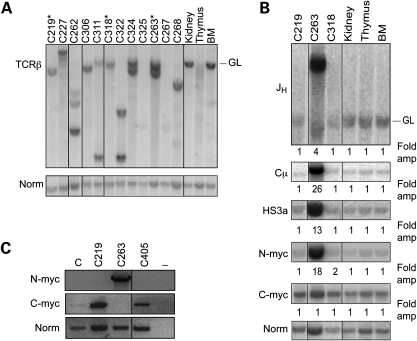
Figure 5.
Clonal rearrangements involving recombining loci in Art-P70/p53 tumors. (A) Analysis of TCR rearrangement status in Art-P70/p53 thymic lymphomas. Genomic DNA isolated from ArtP70/P70p53−/− tumors was digested with EcoRI and then analyzed by Southern blotting. Individual tumors are indicated (top). Thymus, kidney and bone marrow (BM), control tissues; GL, unrearranged, germline band. Amounts of input DNA were normalized to a non-rearranging locus (LR8). *, pro-B lymphomas (C219, C263, C318). (B) Analysis of Art-P70/p53 pro-B lymphomas by Southern blotting. Genomic DNA isolated from Art-P70/p53 double-mutant lymphomas was digested with EcoRI and then analyzed by Southern blotting. Previously characterized probes that hybridized to the JH, Cµ and HS3A regions of the IgH locus, N-myc on chr. 12 and c-myc on chr. 15 were used, as indicated. Individual tumors are indicated, top; Thymus, kidney, and bone marrow (BM), control tissues; GL, unrearranged, germline band. Amounts of input DNA were normalized to a non-rearranging locus (LR8). Fold amplification compared with the non-rearranging locus was calculated as described in Materials and methods. (C) Semi-quantitative RT-PCR of N-myc and c-myc transcript levels. Total RNA was isolated from primary tumor cells, and RT-PCR was performed. cDNAs were PCR amplified using N-myc (exons 2 and 3), c-myc (exons 1–3) and tubulin-specific primers. Tumor numbers, as indicated; C, total RNA from normal, wild-type LN; −, no-RT control. ArtP70/P70p53−/− tumors: thymic lymphomas: C227, C262, C306, C311, C322, C324, C325, C267, C268; pro-B lymphomas: C219, C263, C318. Art−/−p53−/−; pro-B lymphoma: C405.