Table 1.
Protein alternations induced by stress (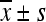 , n = 3)
, n = 3)
| Spot | Protein name | PI | MW | Norm. vol. | |
|---|---|---|---|---|---|
| Control | Stress | ||||
| 1 | Cardiac myosin heavy chain | 7.67 | 76.19 | 0.75 ± 0.24 | 1.54 ± 0.32* |
| 2 | Dihydrolipoamide succinyltransferase component (E2) (E2K) | 7.27 | 62.40 | 0.16 ± 0.08 | 0.57 ± 0.12** |
| 3 | Similar to dihydrolipoamide S-succinyltransferase (E2 component) | 7.09 | 62.64 | 0.32 ± 0.11 | 0.50 ± 0.21* |
| 4 | Mitochondrial aldehyde dehydrogenase | 7.09 | 61.22 | 0.14 ± 0.09 | 0.36 ± 0.10** |
| 5 | Mitochondrial aconitase (nuclear aco2 gene) | 6.93 | 55.61 | 1.29 ± 0.21 | 0.55 ± 0.23* |
| 6 | Albumin | 7.46 | 56.05 | 0.31 ± 0.10 | 1.04 ± 0.21** |
| 7 | Myosin heavy chain, polypeptide 6 | 8.06 | 44.90 | 0.86 ± 0.23 | 1.68 ± 0.48* |
| 8 | Apolipoprotein A-I precursor (Apo-AI) | 7.53 | 30.30 | 0.65 ± 0.22 | 1.39 ± 0.35* |
| 9 | H(+)-transporting ATP synthase | 6.70 | 26.94 | 0.97 ± 0.32 | 1.38 ± 0.46* |
| 10 | Uncoupling protein 3 | 4.35 | 19.02 | 2.15 ± 0.89 | 0.25 ± 0.12** |
*P < 0.05, **P < 0.01 vs control
