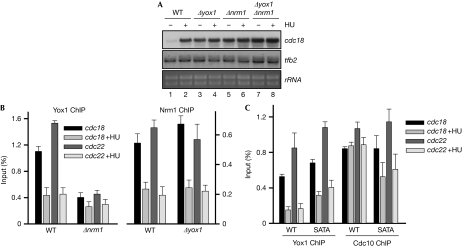
Figure 3.
Nrm1 loads Yox1 onto MBF-dependent genes. (A) Total RNA was prepared from untreated (−) or HU-treated (+) cultures of wild-type (wt) and Δyox1, Δnrm1 and Δyox1 Δnrm1 cells, and analysed by hybridization to the probes indicated on the right. tfb2 and rRNA are shown as loading control. (B) Loading of Yox1 (left panel) or Nrm1 (right panel) on cdc22 and cdc18 promoters was measured by ChIP in untreated or HU-treated (+HU) cultures of wt Δnrm1 and Δyox1 cells. The average of four experiments (±s.d.) is plotted. (C) Loading of Yox1 or Cdc10 on cdc22 and cdc18 promoters was measured in untreated or HU-treated (+HU) cultures of wt and Yox1.SATA (SATA) cells by ChIP. The average of three experiments (±s.d.) is plotted. ChIP, chromatin immunoprecipitation; HU, hydroxyurea; MBF, Mlu1 binding factor.