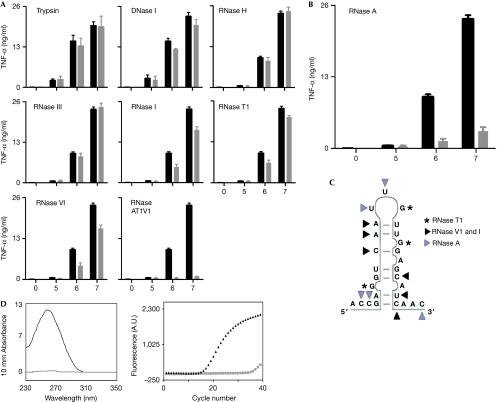
Figure 1.
GBS ssRNA with 3′ of single-stranded cytosine and uracil residues is essential for transcriptional activation of inflammatory cytokines. BMDM from wild-type mice were stimulated with GBS (105, 106 and 107 per ml), which had been depleted for specific macromolecules by enzymatic treatment (grey bars). (A) Trypsin (digestion of proteins), DNase I (degradation of DNA), RNase H (degradation of RNA strands in RNA–DNA hybrids), RNase III (degradation of dsRNA), RNase I (degradation of ssRNA at all dinucleotide pairs), RNase T1 (degradation of ssRNA at 3′ of all single-stranded guanine residues), RNase V1 (degradation of ssRNA at all base-paired nucleotides), combination of RNases A, T1 and V1 (maximal degradation of ssRNAs). (B) RNase A (degradation of ssRNA at 3′ of single-stranded cytosine and uracil residues). (C) Model for sequence and structural specificities of ssRNA-degrading enzyme RNase A (grey triangle), RNase V1 and I (black triangle), RNase T1(asterisk). (D) Absorbance spectrum of RNA and real-time PCR amplification plot of complementary DNA (GBS caf gene). RNA isolated from RNase A-treated (grey) and -untreated (black) bacteria. Data shown are mean values±s.d. (n=3), representative of at least three independent experiments. BMDM, bone-marrow-derived macrophages; dsRNA, double-stranded RNA; GBS, group B streptococci; ssRNA, single-stranded RNA; TNF, tumour necrosis factor; WT, wild-type.