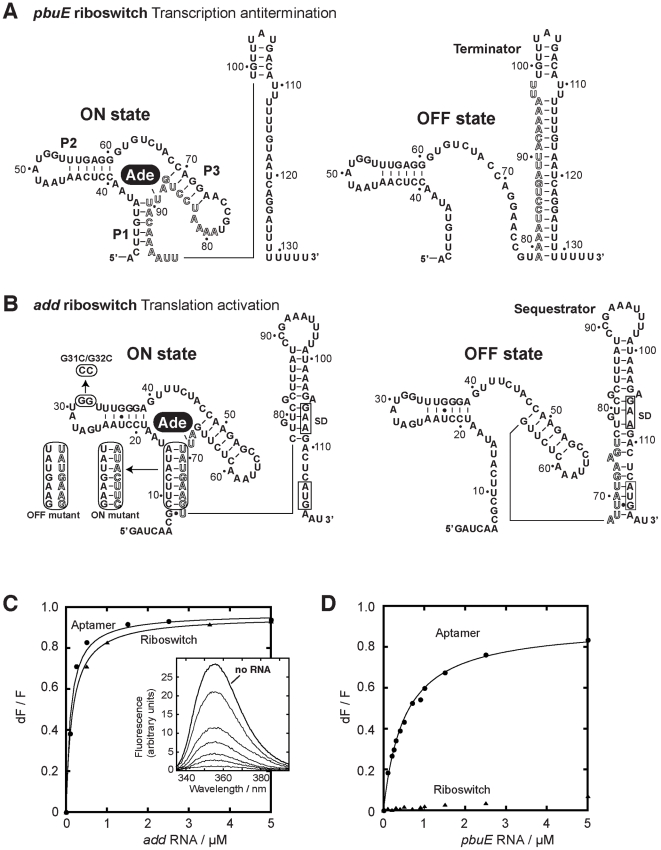
Figure 1. pbuE and add adenine riboswitches exhibit different ligand binding properties.
(A) Secondary structures of the pbuE adenine riboswitch associated with the ON and OFF states. The adenine ligand (Ade) is represented by a black oval. The adoption of the OFF state results in the folding of a terminator structure that promotes premature transcription termination while the presence of adenine favors the ON state and antitermination. Outlined letters represent nucleotides that are involved in the formation of both the terminator and the aptamer structures. The nucleotide numbering is derived from the present study. (B) Secondary structures of the add riboswitch associated with the ON and OFF states. The presence of adenine promotes translational activation. The Shine-Dalgarno (SD) and the AUG initiation codon are both boxed. Mutants used in this study are indicated in rounded rectangles. ON and OFF state mutants are indicated. The P1-3′ mutant corresponds to the wild-type P1 stem but in which the 3′ strand was mutated for the sequence corresponding to the ON state mutant. The nucleotide numbering is derived from this study. (C) Normalized 2AP fluorescence intensity plotted as a function of add aptamer (circles) and riboswitch (triangles) molecules. Changes in fluorescence (dF) were normalized to the maximum fluorescence (F) measured in the absence of RNA. Lines show the best fit to a simple binding model, yielding KDapp of 115±15 nM and 156±7 nM for the aptamer and the riboswitch, respectively. The insert shows fluorescence emission spectra for each add aptamer concentration. The indicated line represents 2AP fluorescence in absence of RNA. (D) Normalized 2AP fluorescence intensity plotted as a function of pbuE aptamer (circles) and riboswitch (triangles) molecules. An apparent binding affinity (KDapp) of 518±27 nM was calculated for the pbuE aptamer. No value was determined for the riboswitch due to the absence of significant fluorescence change.