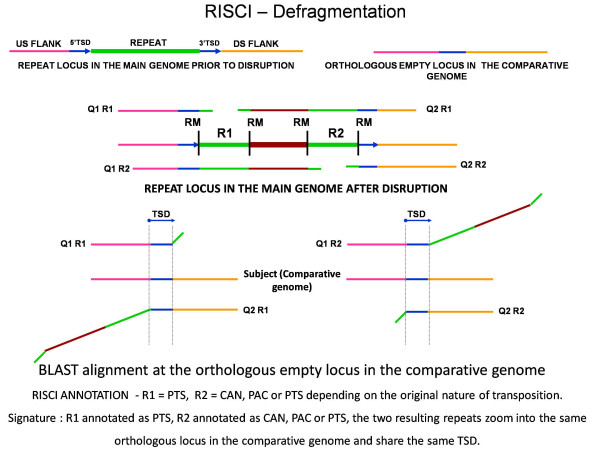
Figure 11.
Alignment signatures for defragmentation by RISCI. The original repeat is disrupted into R1 and R2 due to insertion of an exogenous sequence (brown bold line). Blastn alignments for R1 and R2 at the orthologous empty locus in the comparative genome are shown. Query 2 of R1 (Query2 R1), starting from R1 and downstream, consists of the 50 base overhang into the 3' end of R1, the exogenous sequence (brown line), R2, 3' target site duplication and its downstream flank, of which, only the TSD and its downstream flank find a match at the orthologous empty locus (parallel lines) resulting in PTS annotation for R1. For R2, starting from R2 upstream, Query 1 (Query1 R2) consists of 50 bp overhang into 5' end of R2, the exogenous insertion, R1, 5' TSD and its upstream flank, of which only the TSD and its upstream flank find a match at the orthologous empty locus (parallel lines). RISCI annotation of R2 depends on the nature of the original transposition event and RepeatMasker estimation of the 3' end of repeat (CAN, PAC or PTS). The flank length is crucial in indentifying such events and must be long enough to span the exogenous sequence, R1 or R2 and their respective flanks. From left to right - pink line - 5' flank of the original repeat before disruption, blue arrows and lines - TSDs, R1 - first fragment, brown bold line - exogenous insertion, R2 - second fragment, orange line - 3' flank of original repeat. Query1 R1 - upstream flank of R1 with 50 base overhang into R1, Query2 R1, downstream flank of R1 with 50 base overhang into R1, Query1 R2 - upstream flank of R2 with 50 base overhang into R2, Query2 R2 - downstream flank of R2 with 50 base overhang into R2, TSD - target site duplication sequence.