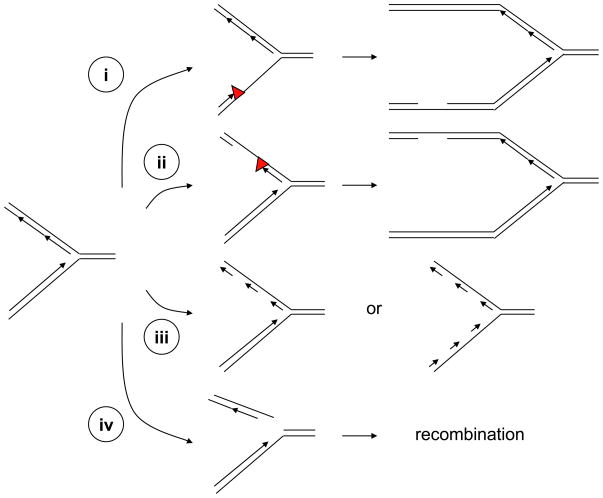
Figure 2. Generation of ssDNA and ssDNA–dsDNA junctions at stressed DNA replication forks.
(i–ii) Stalling of DNA polymerases on either the leading or lagging strand of DNA replication forks can increase the length of ssDNA at the forks. If the lesions (red triangles) are not removed or bypassed, ssDNA gaps can be generated behind the forks. (iii) DNA replication inhibitors such as aphidicolin can inhibit DNA polymerases on the lagging strand or both the leading and lagging strands. Compromised DNA polymerase function leads to increased amounts of ssDNA and an accumulation of short DNA primers at the forks. (iv) DNA lesions such as single-stranded DNA breaks can lead to collapse of DNA replication forks and generate one-ended double-stranded breaks. The resulting DNA breaks can contain ssDNA overhangs and might subsequently be resected by nucleases. The ssDNA generated at stressed replication forks not only contributes to ATR–ATRIP activation, but also participates in the DNA recombination events that facilitate the recovery of stressed forks.