Abstract
Hypermethylation of the MGMT gene promoter and mutation of the TP53 tumor-suppressor gene are frequently present in diffuse astrocytomas. However, there is only anecdotal information about MGMT methylation status and TP53 mutations during progression of low-grade diffuse astrocytoma (AII) to anaplastic astrocytoma (AIII) and secondary glioblastoma (sGB). In this study biopsy specimens from 51 patients with astrocytic tumors with radiologically proved progression from low to high-grade malignancy were investigated for the presence and consistency of MGMT promoter hypermethylation and TP53 mutations. For 27 patients biopsy samples both of primary tumors and their recurrences were available. For the other 24 patients histology of either the low-grade lesion or the high-grade recurrence was available. It was found that MGMT promoter hypermethylation and TP53 mutations are both frequent and early events in the progression of astrocytomas and that their status is consistent over time. No correlation was found between MGMT methylation status and the presence of TP53 mutations. In addition, no correlation was found between MGMT promoter hypermethylation and the type of TP53 mutations. These results argue against the putative TP53 G:C>A:T transition mutations suggested to occur preferentially in MGMT hypermethylated tumors.
Keywords: Diffuse astrocytoma WHO grade II, Anaplastic astrocytoma WHO grade III, TP53 mutation, MGMT promoter hypermethylation, G:C>A:T transition mutations
Introduction
Low-grade diffuse astrocytomas (WHO grade II; AII) are slowly growing tumors with a peak incidence in young adults. They diffusely infiltrate the normal brain and have an intrinsic tendency to progress to greater malignancy, i.e. anaplastic astrocytoma (WHO grade III; AIII) and secondary glioblastoma (WHO grade IV; sGB). Currently available treatment strategies, for example tumor resection, radiotherapy, and/or chemotherapy are only partially effective and most patients develop recurrent or progressive disease. For development of new treatment strategies, identification of genetic alterations associated with pathogenesis, progression, and treatment response of these slow-growing tumors is necessary.
The tumor-suppressor gene TP53 on chromosome 17p13.1 encodes a key transcription factor involved in several cellular mechanisms including growth arrest, DNA repair, and induction of apoptosis. Mutations in the TP53 gene have been reported as early and frequent events in sGB and their precursor lesions [1, 2] whereas de-novo primary glioblastomas reportedly lack TP53 mutations or acquire them late in tumorigenesis [3, 4]. However, the presence of TP53 mutations in the progression of AII to AIII and sGB has mostly been determined in investigations of series of astrocytic tumors with different grades of malignancy without following individual tumors for their tumorigenesis [5, 6]. In some but not all studies on AII, the TP53 mutation was found to be an independent unfavorable predictor of survival and/or malignant transformation [3, 7–9]. Other genetic aberrations in the TP53 pathway, for example MDM2 amplification and p14 ARF deletion or hypermethylation are also frequently observed in low-grade diffuse astrocytomas [7, 10, 11].
Over the last decade, epigenetic silencing of the O6-methylguanine-methyltransferase (MGMT) gene by promoter hypermethylation has been found to be associated with chemosensitivity of a variety of tumor types, including gliomas [12–16]. The MGMT gene is located on chromosome band 10q26 and encodes for O6-alkylguanine-DNA-alkyltransferase (AGT), a repair enzyme that removes promutagenic alkylating DNA adducts from the O6 position of guanine, an important target of alkylating and methylating agents. Lack of MGMT repair capacity contributes to the genesis and progression of human cancers, because it leads to the accumulation of DNA mutations and chromosomal instability [12, 14, 17]. During chemotherapy with alkylating and methylating agents the primary cell mechanism inducing chemoresistence is the demethylating activity of AGT, which removes the alkyl adducts. The absence of MGMT activity can therefore be used as a predictor of response for patients treated with alkylating chemotherapeutic agents. High-grade gliomas, in particular glioblastomas, with hypermethylated MGMT promoters proved to be more sensitive to chemotherapeutic agents, including temozolomide (TMZ), resulting in an overall survival benefit for these patients [13, 15, 18–20].
Apart from the predictive value, the MGMT promoter hypermethylation status has been associated with shorter progression-free survival in AII [21, 22]. In several reports an association between MGMT promoter hypermethylation and an increased frequency of TP53 mutations, with predominance of G>A or C>T transitions (G:C>A:T transition mutations), especially at CpG sites, has been mentioned [11, 21, 23–25]. Further, in a recent population-based study of glioblastomas a higher frequency of TP53 G:C>A:T transition mutations in tumors with MGMT promoter hypermethylation (25%) than in glioblastomas without MGMT methylation (16%, P = 0.0385) was reported [26]. However, information about MGMT promoter hypermethylation or TP53 mutation during astrocytoma progression is sparse [11, 23]. The objective of this study was to investigate the association of MGMT promoter methylation status and TP53 mutations and to determine the consistency of these molecular characteristics over time, in a series of diffuse astrocytomas and their recurrences.
Materials and methods
Patients and tumor samples
Archival routine formalin-fixed and paraffin-embedded (FFPE) tumor tissues were collected from 51 patients diagnosed with:
an AII at the time of the first operation; or
a histopathology of high-grade astrocytic tumor after a wait and see period of at least 1.5 years and a lesion on MR scan without contrast enhancement (clinically regarded as an original low-grade astrocytic tumor).
Glioma recurrences were diagnosed by contrast enhancement on MR scans, with or without clinical symptoms, and/or histopathology of tissue biopsies. For 27 patients tumor tissue was available both from the first operation and from the operation for the recurrence. For 17 patients only tumor tissue from the first operation was available and for seven patients only tissue from the second surgery was available (Table 1). Hematoxylin and eosin (H&E)-stained tissue sections from all tumors were reviewed by an independent neuropathologist (JMK) in accordance with the latest WHO guidelines for typing and grading [27].
Table 1.
Results from histopathological review and molecular analysis
| Case no. | Sex | Age | P/R | Interval (months) | Intervening treatment | Histology | p53 expression % of cells | Mutation analysis of TP53 | LOH of the TP53 locus b | MGMT status |
|---|---|---|---|---|---|---|---|---|---|---|
| Primary only (n = 17) | ||||||||||
| 1 | M | 39 | P | AII | 80 | Exon 6, c.13419A>G, p.Tyr220Cys | Yes | Hypermethylated | ||
| 2 | F | 29 | P | AII | 60 | Exon 4, c.12243_12260dupTACGGTTTCCGTCTGGGC, p.Tyr107_Gly112dup | No | Hypermethylated | ||
| 3 | F | 39 | P | AIII | 60 | Exon 4, c.12253G>T, p.ArgR110Leu/Exon 8, c.14486C>T, p.Arg273Cys | No | Hypermethylated | ||
| 4 | M | 38 | P | AII | 60 | Exon 5, c.13107T>G, p.Val143Gly | Yes | Hypermethylated | ||
| 5 | F | 40 | P | AII | 30 | Exon 8, c.14486C>T, p.Arg273Cys | Yes | Unmethylated | ||
| 6 | M | 38 | P | AII | 40 | Exon 8, c.14496C>A, p.Ala276Asp | Yes | Hypermethylated | ||
| 7 | M | 47 | P | AII | 30 | Exon 8, c.14470_14475dupAACAGC, p.Asn268_Ser269dup | No | Hypermethylated | ||
| 8 | M | 51 | P | AII | Neg. | Exon 5, c.13229delG, p.Asp184 fs | No | Hypermethylated | ||
| 9 | F | 33 | P | AII | 30 | Exon 8, c.14486C>T, p.Arg273Cys | Yes | Unmethylated | ||
| 10 | M | 32 | P | AIII | 20 | No data | Yes | Hypermethylated | ||
| 11 | F | 34 | P | AII | 70 | No data | Yes | No data | ||
| 12 | M | 36 | P | AII | 20 | Exon 8, c.14487G>A, p.Arg273His | No | Hypermethylated | ||
| 13 | F | 59 | P | AII | 60 | Wt | Yes | Unmethylated | ||
| 14 | M | 25 | P | AII | >10 | Exon 5, c.13203G>A, p.Arg175His/Exon 8, c.14513C>T, p.Arg282Trp | No | Hypermethylated | ||
| 15 | M | 38 | P | AIII | 80 | Exon 6, c.13329C>G, p.Pro190Arg | Yes | Hypermethylated | ||
| 16 | M | 43 | P | AII | 80 | Wt | Yes | Hypermethylated | ||
| 17 | M | 39 | P | AII | 50 | Exon 7, c.14061G>C, p.Gly245Ala | Yes | Unmethylated | ||
| Primary and recurrence (n = 27) | ||||||||||
| 18a | M | 35 | P | 4 | RT | AIII | 30 | Exon 5, c.13107T>G, p.Val143Gly | Yes (reverse loss) | Hypermethylated |
| R | GBM | 40 | Wt | Unmethylated | ||||||
| 19 | F | 49 | P | 8 | RT | AIII | 40 | Exon 4, c.12253G>C, p.ArgR110Pro/Exon 5, c.13152G>A, p.Arg158His | No | Hypermethylated |
| R | GBM | 30 | Exon 4, c.12253G>C, p.ArgR110Pro/Exon 5, c.13152G>A, p.Arg158His | No | Hypermethylated | |||||
| 20 | M | 41 | P | 12 | RT | GBM | >80 | Exon 5, c.13152G>C, p.Arg158Pro | No | Unmethylated |
| R | GBM | >80 | Exon 5, c.13152G>C, p.Arg158Pro | Yes | Unmethylated | |||||
| 21 | F | 48 | P | 12 | RT + PCV | AII | <10 | Wt | No | Hypermethylated |
| R | GBM | 80 | Wt | No | Hypermethylated | |||||
| 22 | M | 34 | P | 13 | RT + PCV | AII | 20 | Exon 5, c.13149T>G, p.Val157Gly | Yes | Unmethylated |
| R | AII | 70 | Exon 5, c.13149T>G, p.Val157Gly | Yes | Unmethylated | |||||
| 23 | F | 56 | P | 17 | RT | GBM | >80 | Exon 5, c.13203G>A, p.Arg175His | No | Unmethylated |
| R | GBM | >80 | Exon 5, c.13203G>A, p.Arg175His | No | Unmethylated | |||||
| 24 | F | 36 | P | 18 | RT | AII | 60 | No data | Yes | No data |
| R | AII | 60 | No data | Yes | No data | |||||
| 25 | M | 54 | P | 23 | RT | AIII | >80 | Exon 5, c.13203G>A, p.Arg175His | Yes | Hypermethylated |
| R | GBM | >90 | Exon 5, c.13203G>A, p.Arg175His | Yes | Hypermethylated | |||||
| 26 | F | 31 | P | 23 | RT | AII | >80 | Wt | Yes | Hypermethylated |
| R | GBM | >80 | Wt | Yes | Hypermethylated | |||||
| 27 | F | 55 | P | 25 | RT | AII | <10 | Wt | No | Unmethylated |
| R | AII | <10 | Wt | No | Unmethylated | |||||
| 28 | M | 44 | P | 25 | RT | AII | >10 | Exon 7, c.14057G>T, p.Gly244Cys | Yes | Unmethylated |
| R | AIII | >80 | Exon 7, c.14057G>T, p.Gly244Cys | Yes | Unmethylated | |||||
| 29 | M | 31 | P | 26 | RT + PCV | AII | 20 | Exon 5, c.13107T>G, p.Val143Gly | Yes | Hypermethylated |
| R | OIII | >90 | Exon 5, c.13107T>G, p.Val143Gly | Yes | Hypermethylated | |||||
| 30 | F | 34 | P | 37 | RT | OAIII c | 70 | Exon 6, c.13419A>G, p.Tyr220Cys/Exon 8, c.14487G>A, p.Arg273His | No | Unmethylated |
| R | OAII c | 60 | Exon 6, c.13419A>G, p.Tyr220Cys/Exon 8, c.14487G>A, p.Arg273His | No | Unmethylated | |||||
| 31 | M | 28 | P | 37 | RT | AII | 30 | Exon 6, c.13419A>G, p.Tyr220Cys | Yes | No data |
| R | AIII | 80 | Exon 6, c.13419A>G, p.Tyr220Cys | Yes | Unmethylated | |||||
| 32 | M | 55 | P | 40 | RT + PCV | AII | Neg. | Exon 5, c.13157_13161delATGGC, p.Met160 fs | Yes | Unmethylated |
| R | GBM | Neg. | Exon 5, c.13157_13161delATGGC, p.Met160 fs | Yes | Unmethylated | |||||
| 33 | M | 36 | P | 43 | RT | AII | 30 | Exon 8, c.14487G>A, p.Arg273His | Yes | Hypermethylated |
| R | AIII | >80 | Exon 8, c.14487G>A, p.Arg273His | Yes | Unmethylated | |||||
| 34a | F | 29 | P | 43 | RT | AII | 20 | Exon 7, c.14063A>G, p.Met246Val | Yes | Unmethylated |
| R | GBM | 60 | Exon 8, c.14483G>T, p.Val272Leu | Yes | Hypermethylated | |||||
| 35 | F | 34 | P | 47 | RT | AII | 30 | Exon 5, c.13197T>A, p.Val173Glu/Exon 7, c.14070G>A, p.Arg248Gln | No | Hypermethylated |
| R | GBM | >80 | Exon 5, c.13197T>A, p.Val173Glu/Exon 7, c.14070G>A, p.Arg248Gln | No | Hypermethylated | |||||
| 36a | F | 33 | P | 56 | RT | AII | 30 | Wt | Yes (reverse loss) | No data |
| R | GBM | >80 | Exon 6, c.13419A>G, p.Tyr220Cys | Unmethylated | ||||||
| 37 | F | 39 | P | 62 | RT | AIII | 60 | Exon 5, c.13056A>G, p.Tyr126Cys | No | Hypermethylated |
| R | OAIII | 60 | Exon 5, c.13056A>G, p.Tyr126Cys | No | Hypermethylated | |||||
| 38 | M | 39 | P | 73 | RT | AII | >10 | Exon 8, c.14486C>T, p.Arg273Cys | Yes | Unmethylated |
| R | OAIII | 80 | Exon 8, c.14486C>T, p.Arg273Cys | Yes | Hypermethylated | |||||
| 39 | M | 41 | P | 74 | RT | AII | 50 | Exon 8, c.14486C>T, p.Arg273Cys | Yes | Unmethylated |
| R | AII | 80 | Exon 8, c.14486C>T, p.Arg273Cys | Yes | Unmethylated | |||||
| 40 | M | 55 | P | 85 | RT | AII | <10 | Wt | No | Hypermethylated |
| R | GBM | 80 | Wt | Yes | Hypermethylated | |||||
| 41 | F | 37 | P | 95 | RT | AII | 60 | Wt | Yes | Hypermethylated |
| R | GBM | 80 | Wt | Yes | No data | |||||
| 42 | F | 38 | P | 96 | RT | AII | 80 | Exon 6, c.13418T>C, p.Tyr220His | Yes | Hypermethylated |
| R | AII | >10 | Exon 6, c.13418T>C, p.Tyr220His | Yes | Hypermethylated | |||||
| 43 | M | 46 | P | 146 | RT | AII | 30 | Wt | No | Unmethylated |
| R | AII | 30 | Exon 9, c.14719_14735dupAAGAAACCACTGG, p.Glu326 fs | Yes | Unmethylated | |||||
| 44a | M | 37 | P | 160 | RT | AII | 80 | Exon 9, c.14686C>T, p.Pro309Ser | Yes (reverse loss) | Unmethylated |
| R | AII | 60 | Exon 9, c.14710C>T, p.Gln317Stop | Unmethylated | ||||||
| Recurrence only (n = 7) | ||||||||||
| 45 | M | 43 | R | AII | <10 | Wt | No | Unmethylated | ||
| 46 | M | 43 | R | AII | 20 | Wt | No | Hypermethylated | ||
| 47 | M | 27 | R | GBM | >90 | Exon 5, c.13203G>A, p.Arg175His | Yes | Hypermethylated | ||
| 48 | M | 32 | R | AIII | 70 | Exon 8, c.14486C>T, p.Arg273Cys | Yes | Unmethylated | ||
| 49 | M | 49 | R | GBM | >80 | Exon 8, c.14486C>T, p.Arg273Cys | No | Hypermethylated | ||
| 50 | F | 44 | R | AII | 20 | Wt | No | No data | ||
| 51 | F | 37 | R | GBM | >10 | Exon 5, c.13203G>A, p.Arg175His | Yes | No data | ||
Summary of results from 51 patients diagnosed with a recurrent astrocytic tumor with a clinical history of primary diffuse astrocytoma (WHO grade II)
For 27 patients tumor specimens from primary and recurrent tumor were available; for 17 patients only biopsy material from the first operation was available; and for seven patients only material from the second surgery was available. After molecular investigation of the 27 cases with paired primary tumors and recurrences (cases 18–44), four cases with different TP53 mutations and/or LOH pattern were considered as two, clonally independent, entities (cases 18, 34, 36, and 44)
P, primary; R, recurrence; RT, radiotherapy; PCV, procarbazine, lomustine, and vincristine; AII, low-grade astrocytoma WHO grade II; AIII, anaplastic astrocytoma WHO grade III; GBM, glioblastoma multiforme; OIII, anaplastic oligodendroglioma; OAII, oligo-astrocytoma; OAIII, anaplastic oligo-astrocytoma; Wt, wild-type
aCases with two clonally unrelated tumors, on the basis of the molecular results
bPaired tumors both yes: identical LOH pattern
cThe histology of the recurrence seemed to be indicative of lower malignancy grade than the primary tumor; this is attributed to sampling error in the recurrence (case 30)
All patients received radiation therapy after surgery for the primary tumor. Four of the 27 patients (Table 1; cases 21, 22, 29, and 32) were treated with alkylating chemotherapy (procarbazine, lomustine, and vincristine) between the operations for the primary and recurrent tumors. All patients were treated with temozolomide after the operation for the recurrent tumor. By the end of this study all patients had died. The median follow-up time was 65.5 months.
The archival tissue samples were used in accordance with the code of conduct for appropriate secondary use of tissue: “Proper Secondary Use of Human Tissue” established by the Dutch Federation of Medical Scientific Societies (http://www.federa.org).
DNA extraction
H&E-stained sections from the available FFPE tissue blocks were used by the neuropathologist (JMK) to select parts for DNA extraction. DNA was isolated by standard procedures from selected tissue fragments containing a high percentage tumor cells and, when possible, from fragments of normal tissue [28].
Methylation-specific multiplex ligation-dependent probe amplification (MS-MLPA) for MGMT
The MS-MLPA probe mix used (ME011; MRC Holland, Amsterdam, The Netherlands) includes 21 probe sequences containing an HhaI recognition site, which yields information about the methylation status of the target sequences. In addition, eight control probe sequences not affected by the methylation-sensitive restriction enzyme HhaI digestion are used. The probes containing an HhaI recognition site should only generate a signal if the DNA target is methylated and cannot be digested. The probe mix was developed to detect CpG island methylation of six mismatch repair genes and includes three specific probes for semiquantitative hypermethylation detection of the MGMT promoter region. This MS-MLPA assay for MGMT promoter hypermethylation of gliomas was performed as described by Jeuken et al., with minor modifications [29]. For fragment analysis, PCR products were separated by capillary gel electrophoresis (ABI Prism 3130xl; Applied Biosystems, Foster City, CA, USA) and quantified by use of GeneMarker software version 1.7 (SoftGenetics, State College, PA, USA).
MS-MLPA data analysis
The MS-MLPA results were normalized by dividing the peak height for each MGMT probe signal by the mean peak height for the eight control fragments within the same sample. To estimate the fraction of methylated MGMT promoter DNA, normalized values of each MGMT probe of digested DNA samples were divided by normalized values of corresponding undigested DNAs. Methylation analyses were performed in duplicate and the average ratios of both experiments for each probe were calculated. For the promoter hypermethylation detection of MGMT, we used the MGMT 2 probe from the probe mix only. This probe is located within the widely used CpG island region for MGMT promoter hypermethylation detection by methylation-specific PCR (MS-PCR), and hypermethylation of this region has been found to be an independent predictor of the response of gliomas to TMZ [13, 15, 19, 20].
Preliminary comparison of the widely used MS-PCR and the MGMT 2 probe analyses demonstrated concordant results (not shown). Also, in our hands, the MGMT 2 probe also resulted in better assay reproducibility (Spearman’s rho 0.658, P < 0.001) than the two other MGMT probes (MGMT 1 probe Spearman’s rho 0.448; P = 0.032 and MGMT 3 probe Spearman’s rho 0.431; P = 0.040). Cell lines were used as positive (TE-4, OEC33, FLO-1, SW620) or negative (TE-1, TE-5, TE-13, TE-14, SK-GT-2, BE-3, ESO51) controls for evaluation of MGMT promoter methylation. When the MGMT digested/MGMT undigested ratio of probe MGMT 2 is below 0.30 the MGMT promoter fragment is regarded as non-methylated, as described previously for MS-MLPA assays [30, 31].
Mutation analysis of TP53
Tumor samples from each patient were screened for TP53 mutations in exons 4–9, including intron–exon boundaries, by polymerase chain reaction (PCR) followed by bi-directional direct DNA sequencing using the M13-tailed primer method. Intronic primer sets for PCR amplification are shown in Table 2A. PCR products were generated in a 15-μl reaction mixture containing 1.0 μl DNA solution, 10 μmol of each primer, 25 mM MgCl2, 10 mM dNTPs, and 1U Taq polymerase (Promega, Madison, WI, USA). The PCR reaction was performed using a thermocycler (Biometra, Göttingen, Germany) with an initial denaturing step (95°C) for 3 min, followed by 35 cycles consisting of denaturing (95°C) for 30 s, annealing (60°C) for 45 s, and extension (72°C) for 45 s. After the final cycle, an extension period of 10 min at 72°C was performed. The PCR products (5 μl) were digested with 1 μl exonuclease I (ExoSAP-IT; USB, Cleveland, OH, USA) at 37°C for 30 min followed by inactivation of the enzyme at 80°C for 15 min. The PCR products were sequenced on an ABI Prism 3130xl genetic analyzer using the ABI Prism BigDye Terminator v3.1 kit (both from Applied Biosystems). Samples were analyzed by use of Mutation Surveyor software package version 3.2 (SoftGenetics, State College, PA, USA) and compared with the public sequence in GenBank (NM_000546).
Table 2.
Primer sets for TP53 mutation analysis and LOH analysis of the TP53 Locus
| Exon | Size (bp)a | Forward primer | Reverse primer |
|---|---|---|---|
| A Nucleotide sequences of primers used for TP53 mutation analysis | |||
| Exon 4A | 300 | 5′-CTGGTAAGGACAAGGGTTGG-3′ | 5′-GATGACAGGGGCCAGGAG-3′ |
| Exon 4B | 249 | 5′-AGATGAAGCTCCCAGAATGC-3′ | 5′-GATACGGCCAGGCATTGAAG-3′ |
| Exon 5A | 222 | 5′-TGCTGCCGTCTTCCAGTTGC-3′ | 5′-CTCACAACCTCCGTCATGTG-3′ |
| Exon 5B | 197 | 5′-CAGCTGTGGGTTGATTCCAC-3′ | 5′-TGAGGAATCAGAGGCCTG-3′ |
| Exon 6 | 263 | 5′-TCAGATAGCGATGGTGAGCA-3′ | 5′-GCCACTGACAACCACCCTTA-3′ |
| Exon 7 | 215 | 5′-CGCACTGGCCTCATCTTG-3′ | 5′-AGGGGTCAGAGGCAAGCAGA-3′ |
| Exon 8 | 248 | 5′-GGGACAGGTAGGACCTGATTT-3′ | 5′-GCATAACTGCACCCTTGGTC-3′ |
| Exon 9 | 232 | 5′-GGAGACCAAGGGTGCAGTTA-3′ | 5′-CCCCAATTGCAGGTAAAACA-3′ |
| B Nucleotide sequences of primers used for LOH analysis of the TP53 Locus | |||
| D17S520 | 142 | 5′-GGAGAAAGTGATACAAGGGA-3′ | 5′-TAGTTAGATTAATACCCACC-3′ |
| D17S786 | 167 | 5′-TACAGGGATAGGTAGCCGAG-3′ | 5′-GCTGTGAGTCCCAAAAGAGG-3′ |
| D17S1353 | 122 | 5′-ATTCCCACTGCCACTCCTTG-3′ | 5′-CAGCTGAGGGATACTATTCAG-3′ |
| D17S1566 | 189 | 5′-AAAGATCCTTATTGCCACTTTACTG-3′ | 5′-CTCTTACCTTGCTGGTGAGATTG-3′ |
| D17S1866 | 175 | 5′-TGGATTCTGTAGTCCCAGG-3′ | 5′-GGTTCAAAGACAACTCCCC-3′ |
aAmplicon sizes based on the UCSC Genome Browser (http://genome.ucsc.edu)
Loss of heterozygosity (LOH) analysis of the TP53 locus (17p13)
Five highly polymorphic microsatellite markers located on chromosome 17p13 (D17S1353, D17S1866, D17S1566, D17S786, and D17S520) were used for LOH analysis. Microsatellites were amplified with fluorescence-labeled forward and unlabeled reverse primers (Table 2B). Because normal DNA from non-neoplastic tissues was not available for most of the tumor samples, allelic losses were determined by analysis of the allelic patterns and by integration of data from multiple highly polymorphic markers, as described elsewhere [32]. LOH analysis was performed using the ABI Prism 3130xl Genetic Analyzer (Applied Biosystems) and the GeneMarker software package version 1.7 (SoftGenetics).
Immunohistochemistry (IHC) for p53
IHC for p53 was performed on 4-μm sections of routine FFPE tissues. Briefly, sections were deparaffinized in xylene and hydrated through descending ethanol gradients. Endogenous peroxidase was inactivated by treatment with 3% H2O2 in phosphate-buffered saline (PBS). Antigen retrieval was accomplished by boiling the sections for 15 min in 10 mmol/l Tris–EDTA, pH 9.0 in a microwave oven. The sections were incubated for 1 h at room temperature with mouse monoclonal antibody DO-7 against p53 protein (Dako, Glostrup, Denmark) at 1:50 dilution. After rinsing in PBS, immunoreactivity was visualized using the Envision kit (Dako). The sections were subsequently counterstained with Mayer hematoxylin. Although the DO-7 antibody binds to both normal and mutant p53 protein, in general, normal levels of wild-type p53 protein are too low to be detected by immunohistochemistry. In each section, the number of clearly positive cells out of 1000 cells was counted and >10% was regarded as positive [33].
Statistical analysis
Correlation coefficients were calculated by use of Student’s t-test and Fisher’s exact test. Data were regarded as statistically significant at P < 0.05.
Results
The results from histopathological review and molecular analysis are summarized in Table 1. After molecular investigation of the 27 cases of paired primary tumors and recurrences (Table 1, cases 18–44) four cases with different TP53 mutations and/or reverse LOH patterns were regarded as two clonally independent entities (cases 18, 34, 36, and 44). The strongest indication of clonally independent entities is the finding of specific molecular aberrations in the primary tumors which are not found in the recurrences. In case 43 neither a TP53 mutation nor LOH was found in the primary tumor whereas in the recurrence a TP53 mutation and LOH were detected. This could indicate clonally independent tumors. However, because no molecular aberrations were detected in the primary tumor and because of the long period of time between occurrence of the primary lesion and recurrence (146 months) we regard case 43 as more likely to be clonally related tumors with generation and clonal selection of the aberrations during the long time interval between both tumors.
Patients and tumor histology
According to central review, 34 of 44 (77%) primary tumors were diffuse astrocytomas of WHO grade II (AII); 6 of 44 (14%) were anaplastic astrocytomas (AIII), and 3 of 44 (7%) were glioblastoma (GB). One primary tumor was diagnosed as an anaplastic mixed oligoastrocytoma (OAIII). Of the recurrent tumors 10 of 34 (29%) were still AII; 5 of 34 (15%) had progressed to AIII; and 15 of 34 (44%) became secondary glioblastomas (sGBs). Two recurrent tumors were diagnosed as anaplastic mixed oligoastrocytomas (OAIII), one as anaplastic oligodendroglioma (OIII), and one as low-grade oligoastrocytoma (OAII). In all tumors with an oligodendroglial component found by central review, loss of chromosome 1p and/or 19q was excluded in these cases (results not shown).
Of the 27 cases of which both the primary and recurrent tumor were investigated, 23 cases were regarded as having clonally related tumors and four cases clonally unrelated tumors, on the basis of molecular results. Thirteen of 23 (57%) clonally related tumor pairs showed histological progression of malignancy grade. In nine cases (39%) the tumor histology had not progressed in malignancy grade. In one case the histology of the recurrence seemed to indicate lower malignancy grade than the primary tumor; this was attributed to sampling error in the recurrence (Table 1, case 30). For the 23 paired, clonally related histological samples the mean interval between surgery for the primary tumor and recurrence was 45.1 ± 35.2 months (range 8–146 months). The mean interval between surgery for primary and recurrent tumors of those cases in which the histology of the first biopsy had not progressed in malignancy grade was 50.0 months; for the tumors in which histological progression had occurred the mean interval was 41.3 months (Student’s t-test, P = 0.57).
Hypermethylation analysis of the MGMT promoter
Seventy-eight tumor samples were available for MGMT promoter hypermethylation and TP53 mutation analysis (viz.: 21 samples from patients for whom only the first specimens were available; 54 samples from 27 patients for whom both the first and second surgery tissue samples were available; and three samples from patients for whom only the second tumor specimens were available). Results showing MGMT promoter hypermethylation by MS-MLPA were obtained for 70 of 78 (90%) of the (routine FFPE) tumor tissues (Table 1). MGMT promoter hypermethylation was identified in 39 of these 70 (56%) tumor samples. There was no relationship between tumor grade (viz., AII, AIII or sGB) and methylation status (53% vs. 64% vs. 56% hypermethylated, respectively). For 20 of 23 clonally related tumor pairs MGMT promoter hypermethylation results were obtained from both tumors. In 18 of 20 cases (90%), primary and recurrent tumors had a similar MGMT promoter methylation status (nine tumor pairs hypermethylated; nine tumor pairs not methylated). For two pairs of related tumors a change of MGMT methylation status between the primary and the recurrent tumors was found. There was no relationship between change in MGMT methylation status and intervening treatment with alkylating chemotherapy. Of the four cases with clonally independent tumors, one had different MGMT hypermethylation status, two had identical MGMT hypermethylation status, and for one case no data could be obtained from the second tumor.
TP53 mutation analysis and p53 expression
TP53 exon 4–9 DNA sequence data were obtained from 74 of 78 (95%) tumor samples. Four samples had inferior DNA quality resulting in no reliable sequencing data. TP53 mutation analysis revealed mutations in 40 of the 52 (77%) independent cases. In 35 of these 40 (88%) independent cases one mutation, and in 5 of 40 (13%) cases two TP53 mutations within the same tumor sample were detected. TP53 mutations were detected in 28 of 41 (68%) tumors with histological diagnosis of AII, in 10 of 10 (100%) AIII, and in 13 of 18 (72%) GB (Table 1). Mutation of TP53 was found in 17 of 23 (74%) tumor pairs (primary tumors and their recurrences) regarded as clonally related. In three of these 17 tumor pairs two different TP53 mutations in the same tumor sample were detected and these mutations all remained present in the recurrent tumors. Two tumor pairs had TP53 mutations in the recurrent tumors only, and in one case TP53 mutation was only found in the primary tumor. In two tumor pairs the mutation in the primary tumor differed from that in the recurrence. In five tumor pairs TP53 mutations were not found in either the primary or recurrent tumor, and in one tumor pair no sequence data were obtained.
A total of 45 TP53 mutations were detected in 40 of 55 independent tumors. Three mutations were found in TP53 exon 4, fifteen in exon 5, six in exon 6, four in exon 7, fourteen in exon 8, and three in exon 9. A not previously reported in-frame duplication of 18 nucleotides (codon 107–112) in exon 4 was found in one patient (Table 1, case 2). Of all the TP53 mutations identified in independent cases, 20 of 45 (44%) were G:C>A:T transition mutations; of these, 18 of 20 (90%) were located at CpG sites. Of the 25 not G:C>A:T transition mutations only three mutations (12%) were located at CpG sites. The two G:C>A:T transition mutations that were not located at CpG sites were found in exon 9, the exon of TP53 without CpG sites.
Positive immunohistochemistry for p53 was associated with either mutation of the TP53 gene (Fisher’s exact test, P = 0.02) or LOH of the TP53 locus (Fisher’s exact test, P = 0.02) or both (Fisher’s exact test, P < 0.001). In five of eight tumor samples without p53 expression neither TP53 mutation nor LOH was found. The remaining three samples without p53 expression had TP53 frameshift mutations and, in accordance with this type of mutation, p53 expression was absent.
LOH analysis of the TP53 locus (17p13)
All 78 tumor samples in the study were informative for at least two of five microsatellite markers on chromosome 17p13. In 38 of 48 (78%) tumors with a single TP53 mutation, LOH of the TP53 locus was found. In all tumors in which two TP53 mutations were found, no allelic loss on 17p13 was observed. LOH of the TP53 locus was found in five out of 13 (38%) AII without TP53 mutation and in four out of seven (57%) recurrent tumors without TP53 mutation. The allelic loss pattern was different between three tumor pairs in which LOH was found in the recurrent tumor only. Further, reverse allelic loss of the primary and recurrent tumor was detected in three patients. One of these had a different mutation in each tumor whereas two patients had a mutation in only one of the tumor pairs.
Correlation between MGMT promoter hypermethylation and TP53 mutations
There was no significant correlation between MGMT promoter hypermethylation and TP53 mutation (Fisher’s exact test, P = 0.77; Table 3). In addition, there was no significant correlation between MGMT promoter hypermethylation and the presence of G:C>A:T transition mutations in TP53. G:C>A:T transition mutations were found in 10 of 21 (48%) independent tumors with a hypermethylated MGMT promoter and a TP53 mutation, and in nine of 16 (56%) MGMT promoter unmethylated tumors with a TP53 mutation (Fisher’s exact test, P = 0.80; Table 3).
Table 3.
Correlation between MGMT promoter hypermethylation status and TP53 mutation status
| MGMT hypermethylated (n = 29)a | MGMT not hypermethylated (n = 20) | P value | |
|---|---|---|---|
| TP53 mutated | 21 | 16 | 0.77 |
| G:C>A:T mutation | 10 | 9 | 0.80 |
| G>A transition | 7 | 2 | |
| At CpG site | 7 | 2 | |
| C>T transition | 3 | 7 | |
| At CpG site | 3 | 5 | |
| TP53 not mutated | 7 | 4 |
aCase 10: no data on TP53 mutation could be obtained
Discussion
In this study we demonstrated that MGMT promoter hypermethylation and TP53 aberrations are early and frequent events in the progression of astrocytomas and that both are largely consistent over time. In addition, we did not find a correlation between MGMT promoter methylation status and TP53 mutations. These results argue against the putative TP53 G:C>A:T transition mutations which have been suggested to occur preferentially in MGMT hypermethylated tumors.
MGMT promoter hypermethylation
No data on the MGMT promoter methylation status during astrocytic tumor progression have yet been reported. In this study, we showed that MGMT promoter hypermethylation is not limited to high-grade astrocytomas but should be regarded as an early and frequent event in primary AII and AIII, which remains present during tumor progression. MGMT promoter hypermethylation was found in 58% of primary AII, consistent with frequencies reported by Nakamara et al., Watanabe et al., and Nakasu et al. [11, 23, 34], who demonstrated MGMT promoter hypermethylation by MS-PCR or immunohistochemistry in 48, 63, and 68% of AII, respectively.
Discrepancies between results from immunohistochemistry and from PCR experiments should be taken into consideration. We found concordance in MGMT promoter hypermethylation status between primary and recurrent tumors in 18 of 20 (90%) clonally related tumor pairs. The remaining two cases demonstrate that MGMT promoter hypermethylation may change during astrocytoma progression, e.g., by reversibility of the hypermethylation, clonal selection of methylated or unmethylated subclones, or by loss of the hypermethylated MGMT allele. No relationship between a change in MGMT promoter methylation status and intervening treatment with alkylating chemotherapy was revealed. Recently, Brandes et al. reported their study on the stability of MGMT methylation status in a series of 38 patients with a glioblastoma and second surgery for recurrence. MGMT methylation status changed in 37% of the patients and more frequently in methylated than in unmethylated tumors [35].
TP53 mutations
With regard to the analyses of molecular aberrations in tumors it must be considered that, because of the general procedures used, technically only aberrations present in most of the cells from which the DNA was isolated (i.e. clonal aberrations) will be detected. Tumors are composed of neoplastic and non-neoplastic (normal) cells and it is generally accepted that the neoplastic cells, in contrast with normal cells, harbor clonal molecular aberrations. DNA isolated from a part of the tumor is composed of the DNA of the neoplastic cells and the DNA of the normal cells. These facts imply that when a specific molecular aberration is reliably identified in the DNA sample isolated from the tumor, this DNA sample is retrieved from a part of the tumor composed of a high percentage of clonal neoplastic cells. Admixture of a high percentage of normal cells or the presence of heterogeneous (with regard to the determined aberration) neoplastic cell populations would have masked the aberration and prevented its detection.
The high percentage of mutations of TP53 (77%) detected in the astrocytic tumors in this study is in accordance with the highest frequencies reported in the literature [1, 3, 6–8]. The presence or absence of nuclear p53 expression, determined by immunohistochemistry, was highly significantly associated with the presence or absence of TP53 aberrations. Significant correlation between p53 expression, TP53 mutations, and TP53 locus LOH was shown in other studies [1–3, 8]. In 13 cases with high p53 expression and/or LOH of chromosome 17p13 no TP53 mutations were detected. This may be the result of intratumoral heterogeneity of TP53 gene mutations [36] or TP53 mutations escaping detection by our sequencing method (especially in cases without detected LOH, probably because of admixture of normal DNA). Yet another explanation may be that TP53 mutations were located outside the investigated region of the gene (especially in cases with LOH). In ten cases a single TP53 mutation was detected without concomitant TP53 locus LOH. It may well be that in these cases LOH was probably not identified in the tumor DNA because of too much admixture of normal DNA. In addition, TP53 LOH could be absent because of the presence of a dominant-negative TP53 mutation in the tumors without concomitant loss of the wild type allele. In five cases two TP53 mutations were found and in all eight tumor samples from these cases no LOH of the TP53 locus was observed, which is more supportive of monoclonal tumor cell populations with bi-allelic TP53 mutations than of dual clonality.
TP53 mutations are consistent during astrocytoma progression, because we demonstrate identical TP53 mutation and TP53 locus LOH status in 21 of 27 investigated pairs of primary tumor and recurrence, indicating clonal relationships between these tumor pairs. In two cases TP53 aberrations were found in the tumor recurrences only. It may well be that the mutations were present in the first samples but not detected because the percentage of the mutation-carrying tumor cells was too low. The fact that in both primary tumors no TP53 LOH was observed and only a low percentage of p53 positive cells was seen also indicates the presence of many normal cells in these tumor specimens. In four of 27 (15%) patients differences in TP53 mutation and TP53 LOH between the primary tumors and recurrences were unequivocally found. (One of these cases, case 44, is illustrated in Fig. 1) Allelotyping excluded tissue admixture in these cases. The differences between the primary and recurrent tumors may well be the result of intratumoral heterogeneity or alternatively, may be indicative of two, clonally independent, tumors. Intratumoral heterogeneity for TP53 aberrations has been described previously for astrocytic brain tumors [36] and in the setting of systemic metastasis of glioblastomas [37]. However, in a large study on 144 biopsies from 67 patients with recurrent astrocytoma no indication of intratumoral heterogeneity was obtained [1]. In our study all eight tumor samples from the four paired cases reliably demonstrated TP53 locus LOH with loss of the reverse alleles in three cases, indicating that the isolated DNA was retrieved from a high percentage of clonal tumor cells with little admixture of normal cells or tumor cells of another clone. In addition, in cases 34 and 44 (Table 1) the TP53 mutation identified in the primary tumor was not found in the recurrence and, vice versa, the mutation detected in the recurrence was not present in the primary tumor. Because TP53 mutation is an early event in astrocytoma tumorigenesis the finding of different TP53 mutations in paired primary tumor and recurrence is supportive for dual clonality. Although intratumoral heterogeneity cannot be excluded as the cause of different TP53 aberrations in the paired primary tumors and recurrences, we believe our results are more supportive for the presence of two, clonally unrelated, entities in these four patients.
Fig. 1.
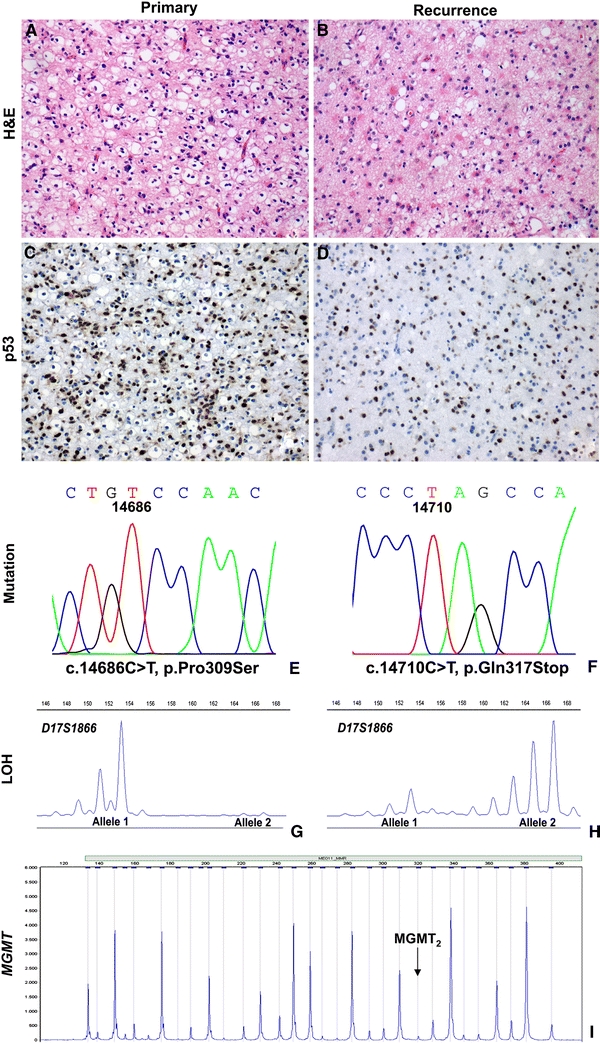
Combined analysis of TP53 and MGMT, case 44 primary and second tumor. a Diffuse astrocytoma (WHO grade II); primary tumor (the perinuclear halos are interpreted as edema of the neuropilema rather than an oligodendroglial tumor component). b Diffuse astrocytoma (WHO grade II); second tumor. c, d Immunohistochemistry for p53 showing many positive (brown) nuclei in both tumor samples. e, f TP53 mutation analysis: different mutations in the primary and secondary tumors, suggestive of the presence of dual clonality. g Loss of heterozygosity (LOH) analysis showing loss of the larger allele in the primary tumor (microsatellite marker D17S1866). h LOH analysis showing loss of the shorter allele in the second tumor (reverse loss, indicative of dual clonality). i MGMT promoter hypermethylation analysis (digested sample), both tumors were not hypermethylated for MGMT (MGMT digested/MGMT undigested ratio of MGMT 2 probe <0.30)
Thirteen of the 23 clonally related pairs of primary tumor and recurrence demonstrated histologically confirmed malignant progression and all cases were consistent with regard to TP53 aberrations (mutation, LOH and p53 expression). This result is in contrast with a study by Sarkar et al. who reported increased p53 expression upon malignant progression of astrocytic tumors. However, in that study only p53 expression was determined and no TP53 mutation nor LOH analysis was performed. Comparable with our study, no correlation between TP53 aberration and interval to recurrence was found [38].
Correlation between MGMT promoter hypermethylation and TP53 mutation
Methylation of the MGMT promoter, and thus absence of AGT expression in gliomas reportedly correlates with G:C>A:T transition mutations of TP53, in particular at CpG sites [11, 21, 23–25]. The mechanism of the preferential occurrence of these mutations may be related to the cytosine methylation-enhanced formation of adducts at methylated CpG sites. A 5-methylcytosine adjacent to the 5′ position of an O6-methylated guanine strongly hampers repair of this defect by AGT. Accordingly, inactivation of MGMT gene expression causes retention of the methylated adducts at the O6 position of guanine. This results in spontaneous or factor-mediated deamination which converts the 5-methylcytosine into thymine. During DNA replication thymine is incorporated and subsequently a G:C to A:T transition mutation may originate at that spot. This would support the relationship between epigenetic inactivation of MGMT and accumulation of this mutation in TP53 at CpG sites [23, 26].
In this study 90% of the G:C>A:T transition mutations were located at CpG sites. However, we neither found a significant correlation between MGMT hypermethylation and TP53 mutation, nor a correlation between MGMT hypermethylation and specific TP53 G:C>A:T transition mutations. Zawlik et al. found a significantly higher frequency of TP53 G:C>A:T transition mutations in MGMT promoter hypermethylated glioblastomas, whereas the total frequency of TP53 mutations in MGMT promoter hypermethylated and unmethylated glioblastomas was similar. Interestingly, no association of MGMT promoter hypermethylation and the accumulation of G:C>A:T transition mutations in PTEN or other genes was found in that study [26], suggesting specific involvement of the TP53 gene. However, in colorectal tumorigenesis and gastric cancer inactivation of MGMT by promoter hypermethylation seemed to be associated with G>A mutations in K-ras [39–41]. In accordance with our findings, Jesien-Lewandowicz et al. recently reported no correlation between methylation of the MGMT promoter and G:C>A:T TP53 mutations in a series of 32 primary glioblastomas treated with radiotherapy and surgery [42]. Future studies may reveal whether the specific methylation of MGMT is representative of a generalized status of genomic methylation and whether there is a gene-specificity of the G:C>A:T transition mutations.
Acknowledgements
We thank Ludo Uytdewilligen for excellent technical assistance and Frank van der Panne for assistance with the photographic work.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Johan M. Kros and Winand N. M. Dinjens contributed equally to this study.
References
- 1.Watanabe K, Sato K, Biernat W, et al. Incidence and timing of p53 mutations during astrocytoma progression in patients with multiple biopsies. Clin Cancer Res. 1997;3:523–530. [PubMed] [Google Scholar]
- 2.Reifenberger J, Ring GU, Gies U, et al. Analysis of p53 mutation and epidermal growth factor receptor amplification in recurrent gliomas with malignant progression. J Neuropathol Exp Neurol. 1996;55:822–831. doi: 10.1097/00005072-199607000-00007. [DOI] [PubMed] [Google Scholar]
- 3.Peraud A, Kreth FW, Wiestler OD, et al. Prognostic impact of TP53 mutations and P53 protein overexpression in supratentorial WHO grade II astrocytomas and oligoastrocytomas. Clin Cancer Res. 2002;8:1117–1124. [PubMed] [Google Scholar]
- 4.Ohgaki H, Kleihues P. Genetic pathways to primary and secondary glioblastoma. Am J Pathol. 2007;170:1445–1453. doi: 10.2353/ajpath.2007.070011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ohgaki H, Dessen P, Jourde B, et al. Genetic pathways to glioblastoma: a population-based study. Cancer Res. 2004;64:6892–6899. doi: 10.1158/0008-5472.CAN-04-1337. [DOI] [PubMed] [Google Scholar]
- 6.Ohgaki H, Kleihues P. Population-based studies on incidence, survival rates, and genetic alterations in astrocytic and oligodendroglial gliomas. J Neuropathol Exp Neurol. 2005;64:479–489. doi: 10.1093/jnen/64.6.479. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe T, Katayama Y, Yoshino A, et al. Deregulation of the TP53/p14ARF tumor suppressor pathway in low-grade diffuse astrocytomas and its influence on clinical course. Clin Cancer Res. 2003;9:4884–4890. [PubMed] [Google Scholar]
- 8.Stander M, Peraud A, Leroch B, et al. Prognostic impact of TP53 mutation status for adult patients with supratentorial World Health Organization Grade II astrocytoma or oligoastrocytoma: a long-term analysis. Cancer. 2004;101:1028–1035. doi: 10.1002/cncr.20432. [DOI] [PubMed] [Google Scholar]
- 9.Okamoto Y, Di Patre PL, Burkhard C, et al. Population-based study on incidence, survival rates, and genetic alterations of low-grade diffuse astrocytomas and oligodendrogliomas. Acta Neuropathol. 2004;108:49–56. doi: 10.1007/s00401-004-0861-z. [DOI] [PubMed] [Google Scholar]
- 10.Ichimura K, Bolin MB, Goike HM, et al. Deregulation of the p14ARF/MDM2/p53 pathway is a prerequisite for human astrocytic gliomas with G1-S transition control gene abnormalities. Cancer Res. 2000;60:417–424. [PubMed] [Google Scholar]
- 11.Watanabe T, Katayama Y, Yoshino A, et al. Aberrant hypermethylation of p14ARF and O6-methylguanine-DNA methyltransferase genes in astrocytoma progression. Brain Pathol. 2007;17:5–10. doi: 10.1111/j.1750-3639.2006.00030.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Esteller M, Hamilton SR, Burger PC, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is a common event in primary human neoplasia. Cancer Res. 1999;59:793–797. [PubMed] [Google Scholar]
- 13.Esteller M, Garcia-Foncillas J, Andion E, et al. Inactivation of the DNA-repair gene MGMT and the clinical response of gliomas to alkylating agents. N Engl J Med. 2000;343:1350–1354. doi: 10.1056/NEJM200011093431901. [DOI] [PubMed] [Google Scholar]
- 14.Gerson SL. MGMT: its role in cancer aetiology and cancer therapeutics. Nat Rev Cancer. 2004;4:296–307. doi: 10.1038/nrc1319. [DOI] [PubMed] [Google Scholar]
- 15.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 16.Levin N, Lavon I, Zelikovitsh B, et al. Progressive low-grade oligodendrogliomas: response to temozolomide and correlation between genetic profile and O6-methylguanine DNA methyltransferase protein expression. Cancer. 2006;106:1759–1765. doi: 10.1002/cncr.21809. [DOI] [PubMed] [Google Scholar]
- 17.Esteller M, Herman JG. Generating mutations but providing chemosensitivity: the role of O6-methylguanine DNA methyltransferase in human cancer. Oncogene. 2004;23:1–8. doi: 10.1038/sj.onc.1207316. [DOI] [PubMed] [Google Scholar]
- 18.Friedman HS, McLendon RE, Kerby T, et al. DNA mismatch repair and O6-alkylguanine-DNA alkyltransferase analysis and response to Temodal in newly diagnosed malignant glioma. J Clin Oncol. 1998;16:3851–3857. doi: 10.1200/JCO.1998.16.12.3851. [DOI] [PubMed] [Google Scholar]
- 19.Hegi ME, Diserens AC, Godard S, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10:1871–1874. doi: 10.1158/1078-0432.CCR-03-0384. [DOI] [PubMed] [Google Scholar]
- 20.Paz MF, Yaya-Tur R, Rojas-Marcos I, et al. CpG island hypermethylation of the DNA repair enzyme methyltransferase predicts response to temozolomide in primary gliomas. Clin Cancer Res. 2004;10:4933–4938. doi: 10.1158/1078-0432.CCR-04-0392. [DOI] [PubMed] [Google Scholar]
- 21.Komine C, Watanabe T, Katayama Y, et al. Promoter hypermethylation of the DNA repair gene O6-methylguanine-DNA methyltransferase is an independent predictor of shortened progression free survival in patients with low-grade diffuse astrocytomas. Brain Pathol. 2003;13:176–184. doi: 10.1111/j.1750-3639.2003.tb00017.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Everhard S, Kaloshi G, Criniere E, et al. MGMT methylation: a marker of response to temozolomide in low-grade gliomas. Ann Neurol. 2006;60:740–743. doi: 10.1002/ana.21044. [DOI] [PubMed] [Google Scholar]
- 23.Nakamura M, Watanabe T, Yonekawa Y, et al. Promoter methylation of the DNA repair gene MGMT in astrocytomas is frequently associated with G:C → A:T mutations of the TP53 tumor suppressor gene. Carcinogenesis. 2001;22:1715–1719. doi: 10.1093/carcin/22.10.1715. [DOI] [PubMed] [Google Scholar]
- 24.Bello MJ, Alonso ME, Aminoso C, et al. Hypermethylation of the DNA repair gene MGMT: association with TP53 G:C to A:T transitions in a series of 469 nervous system tumors. Mutat Res. 2004;554:23–32. doi: 10.1016/j.mrfmmm.2004.02.011. [DOI] [PubMed] [Google Scholar]
- 25.Watanabe T, Katayama Y, Komine C, et al. O6-methylguanine-DNA methyltransferase methylation and TP53 mutation in malignant astrocytomas and their relationships with clinical course. Int J Cancer. 2005;113:581–587. doi: 10.1002/ijc.20625. [DOI] [PubMed] [Google Scholar]
- 26.Zawlik I, Vaccarella S, Kita D, et al. Promoter methylation and polymorphisms of the MGMT gene in glioblastomas: a population-based study. Neuroepidemiology. 2008;32:21–29. doi: 10.1159/000170088. [DOI] [PubMed] [Google Scholar]
- 27.Louis DNOH, Wiestler OD, Cavenee WK, editors. WHO classification of tumours of the central nervous system. 4. Lyon: IARC; 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van der Sijp JR, van Meerbeeck JP, Maat AP, et al. Determination of the molecular relationship between multiple tumors within one patient is of clinical importance. J Clin Oncol. 2002;20:1105–1114. doi: 10.1200/JCO.20.4.1105. [DOI] [PubMed] [Google Scholar]
- 29.Jeuken JW, Cornelissen SJ, Vriezen M, et al. MS-MLPA: an attractive alternative laboratory assay for robust, reliable, and semiquantitative detection of MGMT promoter hypermethylation in gliomas. Lab Invest. 2007;87:1055–1065. doi: 10.1038/labinvest.3700664. [DOI] [PubMed] [Google Scholar]
- 30.Worsham MJ, Chen KM, Meduri V, et al. Epigenetic events of disease progression in head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2006;132:668–677. doi: 10.1001/archotol.132.6.668. [DOI] [PubMed] [Google Scholar]
- 31.Chen K, Sawhney R, Khan M, et al. Methylation of multiple genes as diagnostic and therapeutic markers in primary head and neck squamous cell carcinoma. Arch Otolaryngol Head Neck Surg. 2007;133:1131–1138. doi: 10.1001/archotol.133.11.1131. [DOI] [PubMed] [Google Scholar]
- 32.Hatanpaa KJ, Burger PC, Eshleman JR, et al. Molecular diagnosis of oligodendroglioma in paraffin sections. Lab Invest. 2003;83:419–428. doi: 10.1097/01.lab.0000059948.67795.ef. [DOI] [PubMed] [Google Scholar]
- 33.Idbaih A, Boisselier B, Marie Y, et al. TP53 codon 72 polymorphism, p53 expression, and 1p/19q status in oligodendroglial tumors. Cancer Genet Cytogenet. 2007;177:103–107. doi: 10.1016/j.cancergencyto.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Nakasu S, Fukami T, Jito J, et al. Prognostic significance of loss of O6-methylguanine-DNA methyltransferase expression in supratentorial diffuse low-grade astrocytoma. Surg Neurol. 2007;68:603–608. doi: 10.1016/j.surneu.2006.12.053. [DOI] [PubMed] [Google Scholar]
- 35.Brandes AA, Franceschi E, Tosoni a, et al. O6-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro-Oncology. 2010;12:283–288. doi: 10.1093/neuonc/nop050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren ZP, Olofsson T, Qu M, et al. Molecular genetic analysis of p53 intratumoral heterogeneity in human astrocytic brain tumors. J Neuropathol Exp Neurol. 2007;66:944–954. doi: 10.1097/nen.0b013e318156bc05. [DOI] [PubMed] [Google Scholar]
- 37.Park CC, Hartmann C, Folkerth R, et al. Systemic metastasis in glioblastoma may represent the emergence of neoplastic subclones. J Neuropathol Exp Neurol. 2000;59:1044–1050. doi: 10.1093/jnen/59.12.1044. [DOI] [PubMed] [Google Scholar]
- 38.Sarkar C, Ralte AM, Sharma MC, Mehta VS. Recurrent astrocytic tumours––a study of p53 immunoreactivity and malignant progression. Br J Neurosurg. 2002;16:335–342. doi: 10.1080/02688697021000007588. [DOI] [PubMed] [Google Scholar]
- 39.Esteller M, Toyota M, Sanchez-Cespedes M, et al. Inactivation of the DNA repair gene O6-methylguanine-DNA methyltransferase by promoter hypermethylation is associated with G to A mutations in K-ras in colorectal tumorigenesis. Cancer Res. 2000;60:2368–2371. [PubMed] [Google Scholar]
- 40.Whitehall VL, Walsh MD, Young J, et al. Methylation of O-6-methylguanine DNA methyltransferase characterizes a subset of colorectal cancer with low-level DNA microsatellite instability. Cancer Res. 2001;61:827–830. [PubMed] [Google Scholar]
- 41.Park TJ, Han SU, Cho YK, et al. Methylation of O(6)-methylguanine-DNA methyltransferase gene is associated significantly with K-ras mutation, lymph node invasion, tumor staging, and disease free survival in patients with gastric carcinoma. Cancer. 2001;92:2760–2768. doi: 10.1002/1097-0142(20011201)92:11<2760::AID-CNCR10123>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 42.Jesien-Lewandowicz E, Jesionek-Kupnicka D, Zawlik L, et al. High incidence of MGMT promoter methylation in primary glioblastomas without correlation with TP53 gene mutations. Cancer Genet Cytogenet. 2009;188:77–82. doi: 10.1016/j.cancergencyto.2008.09.015. [DOI] [PubMed] [Google Scholar]


