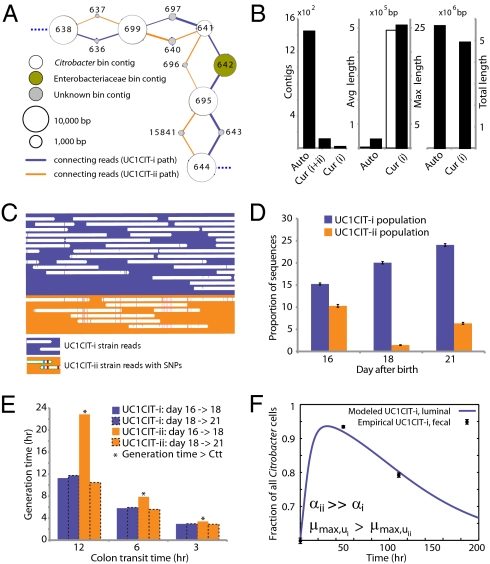
Fig. 3.
Analyses of two ecologically divergent Citrobacter UC1CIT subpopulations. (A) Schematic representation of part of the fragmented UC1CIT assembly. At the ends of many Citrobacter contigs (e.g., contig number 699), reads that partially coassembled led to two sequence paths because they became too divergent (e.g., reads placed in contigs 697 vs. 640) or contained completely novel sequence (reads placed in 642 vs. 696). (B) Condensing of these paths by manual curation reduced the number of contigs, increased average (white bar, left axis) and maximum contig length (black bars, right axis), and reduced cumulative length of contigs (auto, automatic assembly, cur (i+ii), curated UC1CIT-i and -ii strain contigs; cur (i), curated UC1CIT-i contigs). (C) After curation, reads from the strains were grouped based on single nucleotide polymorphism patterns using Strainer. (D) Shifts in proportion (of the respective library total number of reads) of UC1CIT-i and UC1CIT-ii reads over time. Error bar indicates SD between UC1CIT contigs. (E) A chemostat model of the colon, only allowing for differences in growth rate, was used to predict generation time differences needed to explain the observed dynamics in D. (F) Simulation based on a model that incorporated intestinal wall attachment fitted to the observed strain abundances when strain UC1CIT-ii had a higher affinity α to the intestinal wall and the UC1CIT-i luminal maximum growth rate μmax,u was higher than the growth rate of UC1CIT-ii.