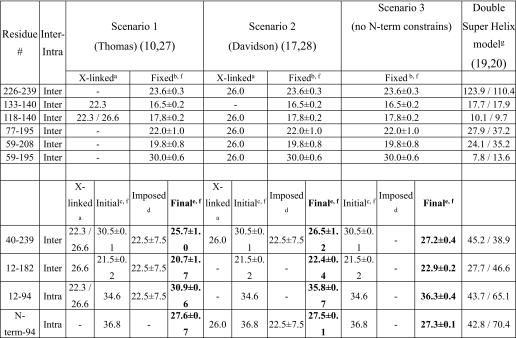
TABLE 2.
Molecular modeling constrains and final distances (Å) of significant residues in the three folding scenarios explored and comparison with double-super helix model predicted distances
a Cross-links were observed by Thomas and co-workers (10, 27) and Davidson and co-workers (17, 28). Cα-lysine-(cross-linker)-Cα-lysine distances depend on the cross-linker used as follows: disuccinimidyl glutarate, 22.3 Å; bis(sulfosuccinimidyl) suberate, 26.0 Å; and dithiobis(succinimidyl propionate), 26.6 Å.
b Cα-lysine-(cross-linker)-Cα-lysine distances from our initial model were derived as described under “Experimental Procedures.” The positions of the corresponding residues (more than residue 50) were fixed during minimization and molecular dynamics of the N-terminal residues (1–49).
c Cα-lysine-(cross-linker)-Cα-lysine distances from our initial model (see under “Experimental Procedures“) were before minimization and molecular dynamics of the N-terminal residues (1–49).
d Imposed distance constraints were during minimization and molecular dynamics of the N-terminal residues (1–49). The distance values were selected based on the experimental cross-links observed by Thomas and co-workers (10, 27) and Davidson and co-workers (17, 28).
e Cα-lysine-(cross-linker)-Cα-lysine final distances after minimization and molecular dynamics of the N-terminal residues (1–49) are shown.
f Averages ± S.D. of two intermolecular distance values for the two monomers of apoA-I are reported.
g The two numbers are distances between the indicated positions within monomer A (intramolecular) or between A–B (intermolecular) and within monomer B (intramolecular) or between B–A (intermolecular), respectively.