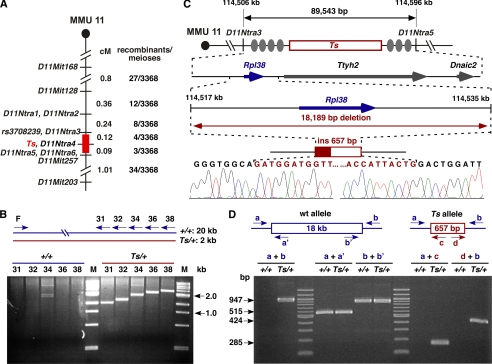
FIGURE 1.
Molecular genetics of Ts. A, genetic map of the Ts interval on distal MMU 11. Shown is the genetic linkage map obtained from the F2 intercross at the distal portion of mouse chromosome 11 (MMU 11), where locations of genetic markers are shown on the right, their genetic distance in centimorgans shown on the left, followed by the number of recombinants per meioses that separate each marker. The nonrecombinant critical Ts region is shown in red. B, insertion/deletion PCR screen. A single forward primer (F) and five progressively more distal located reverse primers (R31, -32, -34, -36, and -38) were predicted to produce products ranging from ∼19.5 to ∼21.0 kb. Instead, products ranging from 1.5 to 3.0 kb were observed exclusively from Ts/+ DNA template, indicating the absence of ∼18 kb of genomic DNA on the Ts chromosome. Shown is a SYBR Green-stained 2% agarose gel separating the amplified products. Lane M, DNA size standard. C, physical map of the Ts critical region. From top to bottom is shown the genetic region on chromosome 11 defined by markers D11Ntra3 and D11Ntra5 with their physical location given at the top in kb. Filled ovals represent the approximate locations of recombination events, and the red box indicates the location of the Ts critical interval. The arrow marks the position and orientation of Rpl38 (ribosomal protein L38), Ttyh2 (tweety homolog 2), and Dnaic2 (dynein, axonemal, intermediate chain 2). Sequencing chromatogram of the PCR fragment amplified from the Ts chromosome. Bases shaded in red represent the insertion sequence. D, verification/genotyping PCR scheme. PCR primers flanking the Ts mutation (a and b), within the deleted region (a′ and b′), and within the insertion (c and d) were generated. All +/+ DNAs tested (n = 9) yielded only fragments a + a′ and b + b′, whereas all Ts/+ DNAs tested (n = 13) yielded a + b, a + a′, b + b′, a + c, and d + b.