Figure 1.
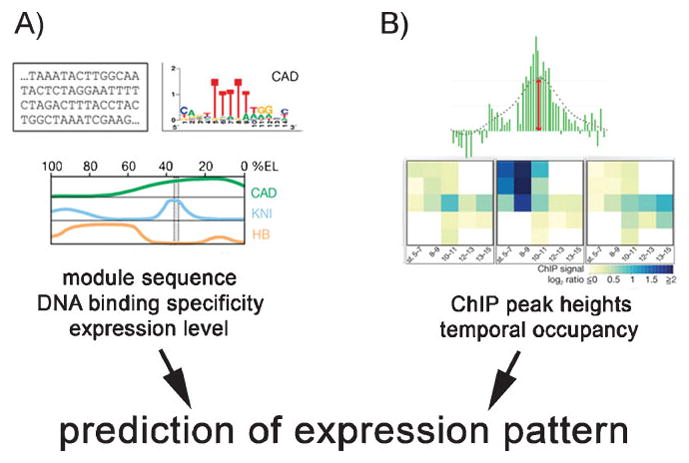
Two computational approaches to predict gene expression patterns differ in their input parameters. A: This approach uses DNA sequence data, TF binding specificity and expression levels. B: This approach is based on the binding intensity of peaks obtained by ChIP-on-chip (see text for details) and temporal occupancy of the regulatory sequence (adapted by permission from Macmillan Publishers Ltd: Nature,[8, 13] copyright 2008, 2009).
