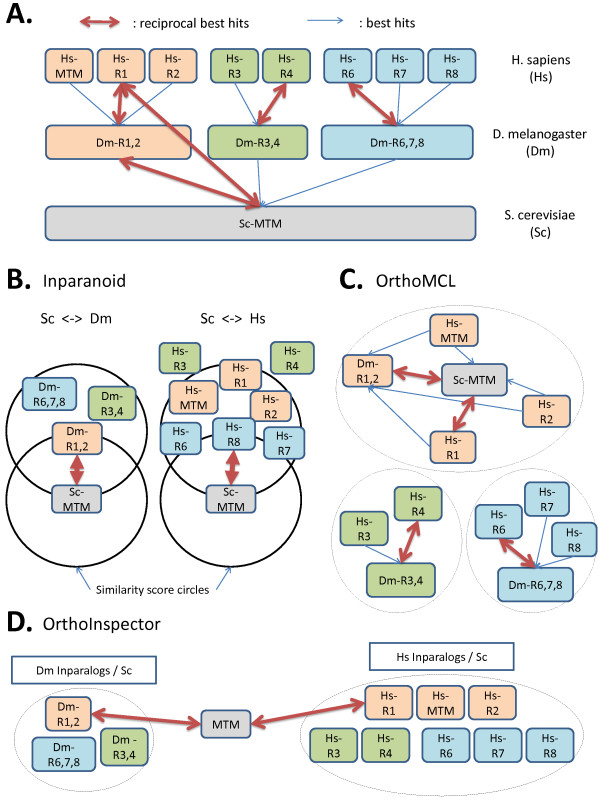
Figure 6.
Myotubularin family predictions. A. The myotubularin family distribution is established in three species: H. sapiens (Hs), D. melanogaster (Dm) and S. cerevisiae (Sc), and multiple duplication events have been identified. Reciprocal best hits (RBH) and best hits (BH) linking the sequences are represented as red and blue arrows. Sequences used are Hs-MTM (MTM_HUMAN), Hs-R1 (MTMR1_HUMAN), Hs-R2 (MTMR2_HUMAN), Hs-R3 (MTMR3_HUMAN), Hs-R4 (MTMR4_HUMAN), Hs-R6 (MTMR6_HUMAN), Hs-R7 (MTMR7_HUMAN), Hs-R8 (MTMR8_HUMAN), Dm-R1,2 (Q9VMI9), Dm-R3,4 (Q7YU03), Dm-R6,7,8 (Q8MLR7), Sc-MTM (P47147). B. Inparanoid first identifies RBHs, then searches for putative inparalogs around these RBHs in a search space delimited by similarity score circles. The prediction between Hs and Sc excludes Hs-R3 and Hs-R4 sequences. C. OrthoMCL constructs a graph with proteins as nodes and similarities as edges, then identifies sub-graphs corresponding to co-ortholog groups. In this example, 5 human myotubularins and 2 fly myotubularins are excluded from the group containing the yeast myotubularin. D. OrthoInspector defines inparalog groups, then compares groups between organisms based on RBH and BH relations. Here, inparalog groups containing all the members of the myotubularin family are identified in human and fly, with no false negatives.