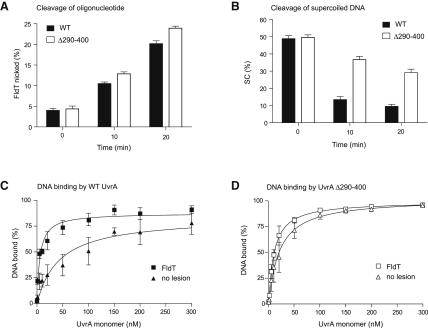
Figure 7.
Activity of UvrA Δ290-400 in Incision and DNA Binding Assays
(A) Nicking of a 50 bp duplex containing a single fluorescein (FldT) adduct. The substrate was incubated at 37°C with UvrA, UvrB, and UvrC, and aliquots were removed and quenched at the times indicated. The samples were analyzed by denaturing acrylamide gel electrophoresis, and the proportion of the radiolabeled DNA fragment that had been nicked was quantified. Data are the mean of three independent experiments and are shown with standard deviation.
(B) Nicking of a UV-irradiated supercoiled plasmid. The plasmid was incubated at 37°C with UvrA, UvrB, and UvrC, and aliquots were removed and quenched at the times indicated. The samples were analyzed by native agarose gel electrophoresis, and nicking was detected as the conversion of supercoiled plasmid to nicked open circle DNA. Data are the mean of three independent experiments and are shown with standard deviation.
(C and D) Binding of UvrA and UvrA Δ290-400 to a 50 bp duplex containing either a single FldT adduct or no lesion. The indicated concentrations of protein were incubated with 1 nM DNA for 20 min at 37°C. Samples were analyzed by EMSA. Data were fitted to a one-site binding equation, and the calculated dissociation constants were as follows: WT UvrA, KdFldt = 5.6 ± 1 nM, Kdno lesion = 43 ± 17 nM and UvrA Δ290-400, KdFldt = 10.5 ± 1 nM and Kdno lesion = 20.8 ± 4 nM. Data are the mean of three independent experiments and are shown with standard error.