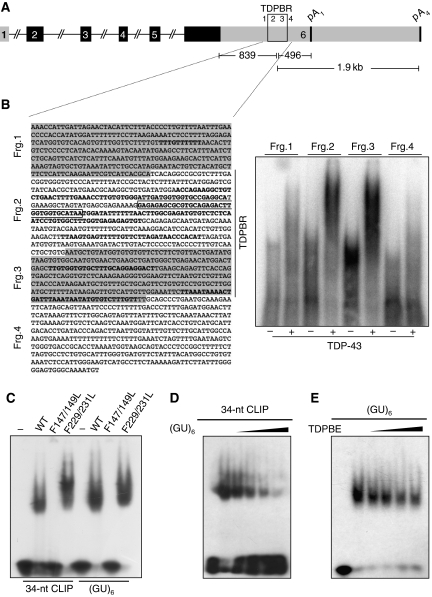
Figure 3.
TDP-43 specifically binds its 3′ UTR region. (A) Schematic representation of the TDP-43 gene depicting the location and sequence of the TDPBR (B, fragments 2 and 3) within the 3′ UTR. Coding exons and untranslated regions are shown in black and grey, respectively. (B) In the left panel, the 3′ UTR region of TDP-43 with the different regions tested for TDP-43 binding (frgs. 1–4) is reported. In this panel, all the CLIP sequences/clusters reported from different experiments (J Ule and J Tollervey unpublished results) are highlighted in bold. The major 34-nt CLIP sequence seen in HEK 293 cells and used in comparative-binding analysis (C–E) is boxed. On the right, band-shift analysis using recombinant TDP-43 using these different fragments. (C) GST-TDP43 mutants were analyzed for binding to the major 34-nt CLIP sequence (boxed) and (GU)6 RNA as control. (D, E) Competition assays using constant levels of wild-type protein to bind radiolabelled (GU)6 as a function of increasing concentrations of unlabelled 34-nt CLIP sequence (D), or binding to radiolabelled 34-nt CLIP sequence as a function of increasing concentrations unlabelled (GU)6. Protein was absent from the first lane in each panel. ∼0.5 μg of the different recombinant proteins were used with 1 ng of the various 5′-labeled single-stranded RNA oligonucleotides. Competition analyses were carried out with 2.5, 3.75, 5, and 10 ng of unlabelled 34-nt CLIP sequence and (GU)6.