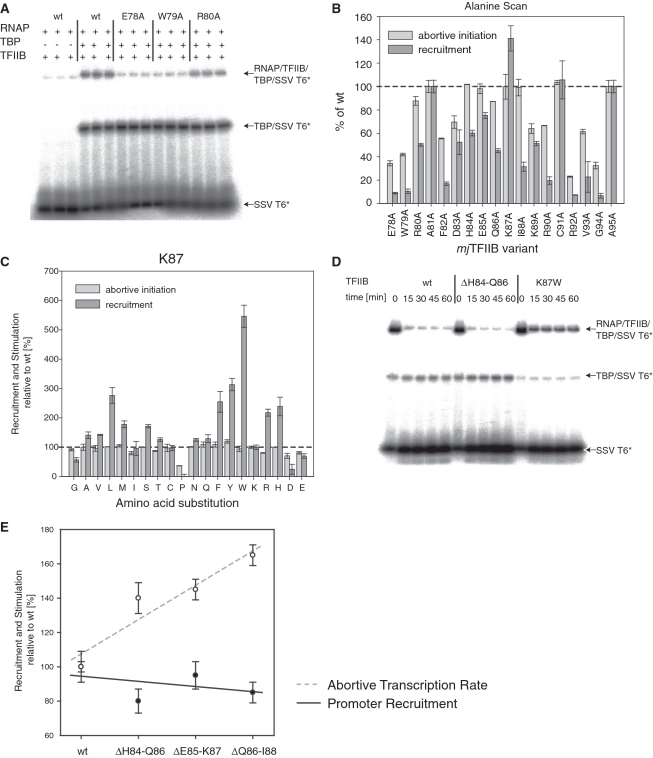
Figure 5.
Recruitment properties of TFIIB linker mutants. (A) Typical EMSA experiment to quantitate the RNAP recruitment activity of mjTFIIB mutants on radiolabelled SSV T6 core promoter (‘SSV T6*’). The arrows identify the various types of complexes formed. (B) Recruitment and stimulatory activities of the mjTFIIB alanine substitution series relative to wild-type mjTFIIB (dotted line). Mutants were tested in triplicate and error bars represent standard deviations. (C) Recruitment and stimulatory activities of the mjTFIIB K87 mutants relative to wild-type mjTFIIB (dotted line). Mutants were tested in triplicate and error bars represent standard deviations. (D) Wild-type TFIIB and TFIIB variants ΔH84-Q86 and K87W were incubated with RNAP, TBP and radiolabelled probe under the usual binding conditions as detailed in ‘Materials and Methods’ section. After 20 min, unlabelled SSV T6 core promoter was added at 400-fold excess as a competitor and samples taken after 0, 15, 30, 45 and 60 min. The addition of competitor DNA reduced the binding of RNAP/TFIIB/TBP to the radiolabelled promoter 5-fold less for mjTFIIB-K87W than for wild-type TFIIB or mjTFIIB ΔH84-Q86. (E) Recruitment (circles) and stimulatory activities (solid dots) of mjTFIIB deletion mutants were calculated relative to wild-type TFIIB (=100%). Mutants were tested in triplicate and error bars represent standard deviations. Multiple regression trendlines for both recruitment activity (black line) and stimulatory activity (dotted grey line) show that increased stimulatory functions of the deletion mutants are distinct from their recruitment activities that remain at or slightly below wild-type levels.