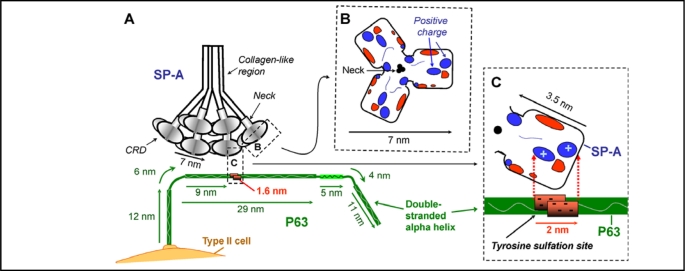
Fig. 7.
Hypothesized interaction of SP-A and P63. Diagrams are drawn approximately to scale. A. The positively charged areas on the surface of the carbohydrate recognition domain (CRD) of SP-A is hypothesized to interact with the double stranded P63 protein at the negatively charged region of P63 predicted to contain a sulfated tyrosine. The length of the P63 luminal C-terminal was drawn as 69 nm, the predicted size of a double stranded helix of 602 amino acids [49]. Structural features of P63 are based on the amino acid composition, the ScanProsite predicted features, and the data of Hauri's group [43, 47, 48, 49] as shown in Fig. 4 and 5. P63 is drawn as a curved rod as seen in EM photomicrographs [49]. R. View of the top of the 7nm long SP-A trimer CRD region, with the location of the electrostatic surface charges shown after calcium binding [18]. Positive charged regions enlarged in diagram C are indicated. C. View of one SP-A molecule in the trimeric CRD of SP-A, estimated to be approx. 3.5 nm in length. In the P63 molecule, the area from 318 to 328 containing 5 negatively charged amino acids is estimated to be approx. 1.6 nm long forming a patch of 2 nm due to the double strands of the coiled coil of P63. The positively charged area on SP-A could interact with the negatively charged tyrosine sulfation site on P63.