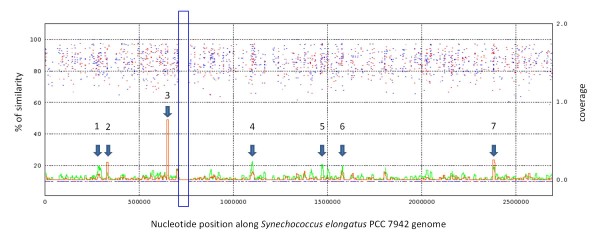
Figure 9.
Coverage of sequences from the metagenomes of two microbialites (Río Mesquitez and Pozas Azules) found in the region of Cuatro Cienegas along the genome of PCC 7942. X axis indicates the nucleotide position along the PCC 7942 genome. Left axis indicates the percent of similarity of aligned metagenomic sequences to the genome of PCC 7942 (blue and red dots correspond to alignments to forward and reverse strands to S. elongatus DNA). The position of these alignments along the genome are also shown at the bottom of the figure to differentiate regions of coverage from non-coverage; right axis indicates a relative measure of coverage of sequences from the metagenomes on the genome of PCC 7942 (the green line at the bottom corresponds to the metagenome from Río Mesquites; while the red line to the metagenome from Pozas Azules). Coverage was calculated for windows of 10,000 nucleotides (sliding 500 nt.) as: (number of aligned nucleotides along the genome)/10,000. The blue rectangle indicates a large region of no coverage along the genome. Larger coverage peaks correspond mostly to genes classified as PX or PHX according to codon usage. The following genes contribute to the largest coverage peaks (1) FtsH peptidase (Synpcc7942_0297, PX); (2) F0F1 ATP synthase subunit alpha (Synpcc7942_0336, PX); (3) photosystem II 44 kDa subunit reaction center protein (Synpcc7942_0656, PHX); (4) ATPase, E1-E2 type (Synpcc7942_1082, PX), glycosyltransferase (Synpcc7942_1083), glycogen branching enzyme (Synpcc7942_1085, PX), ATPase (Synpcc7942_1089, PX); (5) protochlorophyllide reductase iron-sulfur ATP-binding protein (Synpcc7942_1419), light-independent protochlorophyllide reductase subunit N (Synpcc7942_1420, PX), putative carboxysome assembly protein (Synpcc7942_1421); (6) two component transcriptional regulator (Synpcc7942_1453); (7) F0F1 ATP synthase subunit beta (Synpcc7942_2315, PX).