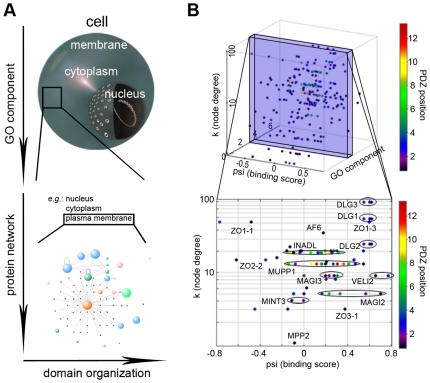
Figure 7. Optimization of PDZ binding selectivity in a four dimensional space.
(A) Schematic illustration of the stepwise mechanism that reduces inherent PDZ binding overlap in the living cell. (B) Practical illustration of the proposed mechanism through interactions with the human plasma membrane protein Neurexin-1. Modeled binding scores are subdivided in cellular space by superimposing gene ontology compartment annotations (numbers as in Fig. 3) and the PDZ protein's node degree. The PDZ position in the protein is color-coded to indicate the presence (e.g. INADL) or absence (e.g. ZO1) of neighboring PDZ domains with similar binding score for (i.e. chance to bind) a given ligand. In the lower panel, PDZ domains of the same protein are encircled to illustrate this. In the cell, having PDZ domains with overlapping binding affinities in a single protein may bring about competition for a specific ligand, but may also increase the overall potential of a PDZ protein to bind the right ligand. Alternatively, splice variations can be envisioned to further specify PDZ selectivity in vivo.