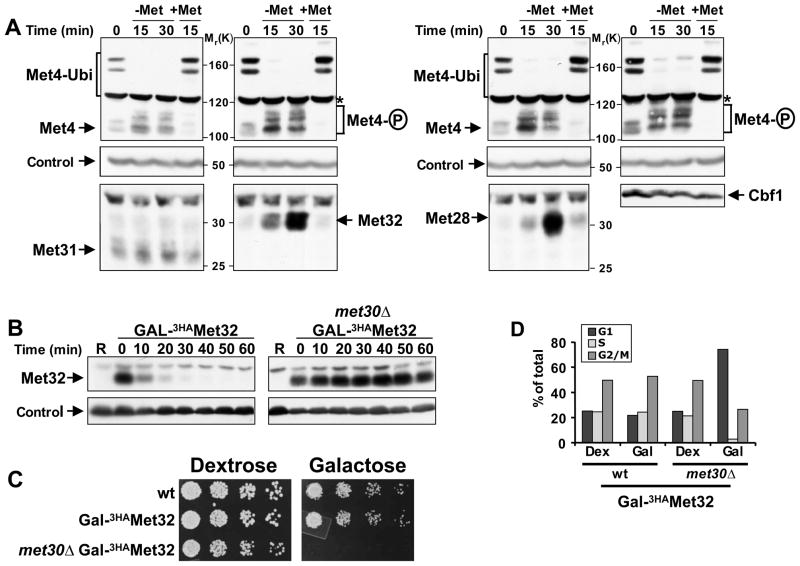
Figure 1. Met32 degradation by SCFMet30 is required for cell proliferation.
(A) Regulation of Met4 transcription complex components. Cells expressing Met313HA, Met323HA, Met283HA, or Cbf13HA from their endogenous loci were grown in minimal medium supplemented with 1mM methionine (+Met), shifted to medium without methionine (−Met) for 30′ to activate Met4, before methionine was added back to the cultures for an additional 15′ (+Met) to inactivate Met4. Cell lysates were analyzed by immunoblotting using anti-HA antibodies (Met28, Met32, Met31, Cbf1), and anti-Met4 antibodies to detect Met4 activation as indicated by loss of the ubiquitylated forms and appearance of phosphorylated species. The proteasome subunit Rpt1 was detected as a loading control.
(B) Met32 degradation depends on the F-box protein Met30. Met32 stability was analyzed by promoter shut-off experiments (“gal-shut-off“) and immunoblotting using anti-HA antibodies. R: uninduced sample grown in raffinose medium.
(C) Degradation of Met32 is essential for cell proliferation. Five fold serial dilutions of yeast strains as indicated were spotted onto YEP-dextrose (no Met32 expression) and YEP-galactose plates (Met32 expression). Plates were incubated at 30° C.
(D) Stabilization of Met32 induces a G1 cell cycle arrest. Yeast cells as indicated were grown in YEP-dextrose or YEP-galactose for 4hrs. Cell cycle distribution was analyzed by flow cytometry.