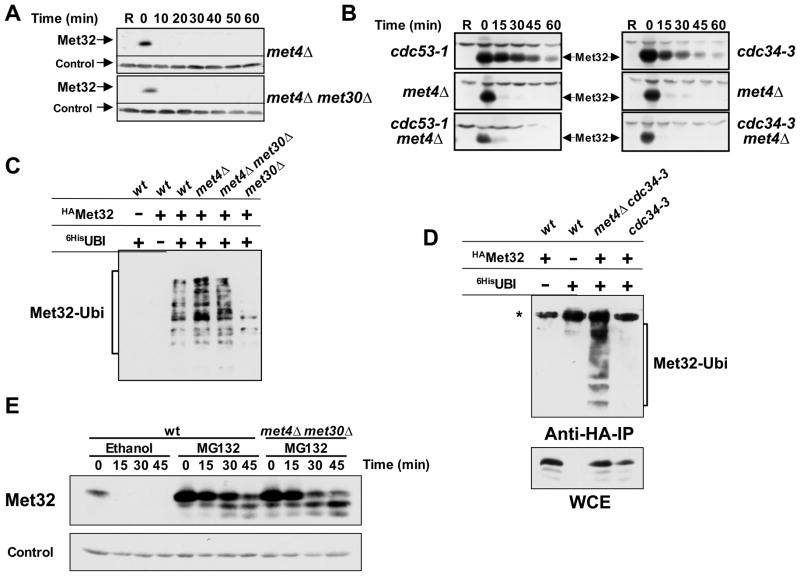
Figure 3. Met32 is degraded by two independent degradation pathways.
(A) Met30-independent degradation of Met32 in the absence of Met4. Met32 stability was assayed as described for figure 1B in met4Δ mutants and met4Δ met30Δ double mutants.
(B) The second degradation pathway of Met32 is independent of Cdc53 and Cdc34. Met32 degradation was analyzed in the mutants indicated as described for figure 1B, except that cells were shifted to non-permissive temperature (35°C) to inactivate Cdc53-1 or Cdc34-3 for 30min prior to the addition of galactose.
(C) Met32 ubiquitylation in vivo. Cells as indicated expressing GAL1-3HAMet32 and CUP1-6xHisubiquitinG76A were cultured in raffinose medium containing 1mM methionine. Expression of 6xHisubiG76A was induced for 2 hrs by addition of 100μM CuSO4, galactose was added for 1 h to induce 3HAMet32 expression, and cells were treated for 45 min with 50μM MG-132 to block proteasome activity. Ubiquitylated proteins were purified under denaturing conditions on Ni2+-sepharose beads and analyzed by immunoblotting using anti-HA antibodies. All strains were pdr5Δ mutants to increase permeability for MG-132 (Fleming et al., 2002).
(D) Cdc34 is required for Met32 ubiquitylation in vivo by the SCFMet30 pathway but not by the second degradation pathway. Experiment as in panel C, but cells were shifted to 35°C 30 minutes before addition of galactose to inactivate Cdc34-3. The asterisk indicates a cross-reacting band that was only visible when cells were grown at high temperature.
(E) Both Met32 degradation pathways are proteasome-dependent. Met32 stability was assayed as described for figure 1B except that cells were treated with 50μM MG-132 or solvent control (ethanol) 20 minutes before adding dextrose.