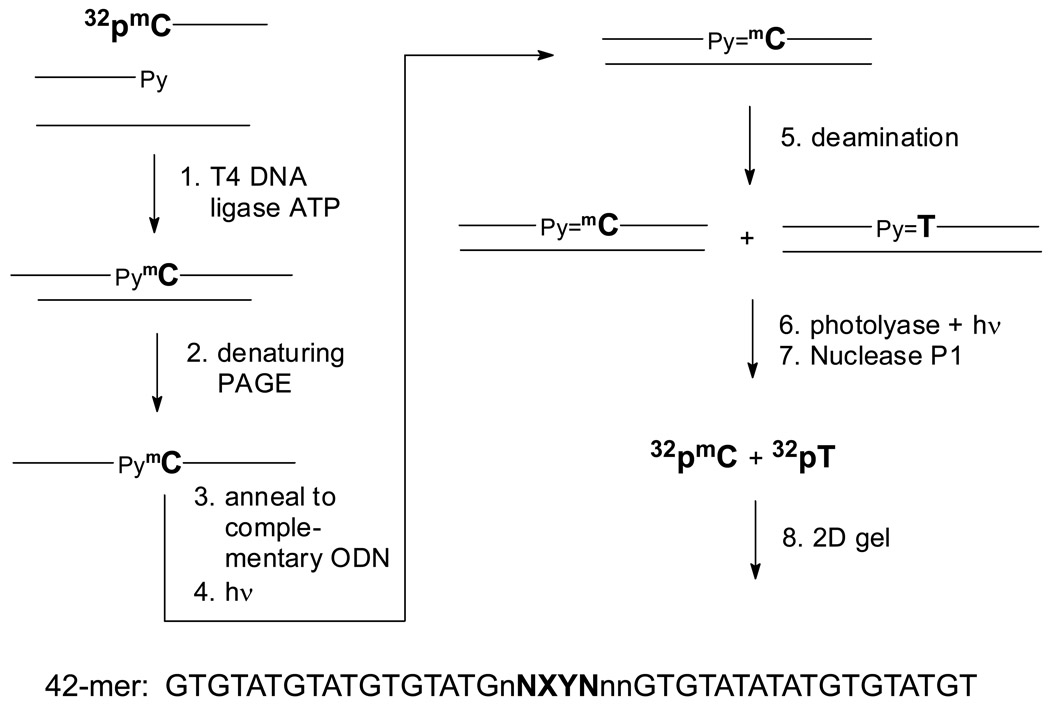
Figure 2.
Scheme for substrate preparation and analysis of the deamination rate of mC-containing cyclobutane dimers in duplex DNA. 1) 5'-32P-labeled 21-mer with a terminal mC is ligated to second 21-mer ODN in the presence of a 20-nt ligation scaffold. 2) The single-strand 42-mer ligation product is separated from unligated products by denaturing gel electrophoresis. 3) The double strand substrate is prepared by annealing the radiolabeled 42-mer substrate to the complementary 42-mer ODN. 4) The duplex is irradiated with 302 nm on ice to generate the CPD. 5) The irradiated product is deaminated for various times at 37°C. 6) The deamination mixture is treated with photolyase and light to photorevert the CPD. 7) Nuclease P1 degrades undamaged and photoreverted DNA to mononucleotides. 8) Two-dimensional acrylamide electrophoresis of the nuclease P1 products separates the non-deaminated mC from its deamination product T.