Abstract
The etiology of a large class of inherited neurological diseases is founded on hairpin structures adopted by repeated DNA sequences, and this folding is determined by base sequence and DNA context. Using single substitutions of adenine with 2-aminopurine, we show that intrastrand folding in repeated CAG trinucleotides is also determined by the number of repeats. This isomeric analog has a fluorescence quantum yield that varies strongly with solvent exposure, thereby distinguishing particular DNA motifs. Prior studies demonstrated that (CAG)8 alone favors a stem-loop hairpin, yet the same sequence adopts an open loop conformation in a three-way junction. This comparison suggests that repeat folding is disrupted by base pairing in the duplex arms and by purine-purine mismatches in repeat stem. However, these perturbations are overcome in longer CAG repeats, as demonstrated by studies of isolated and integrated forms of (CAG)15. The oligonucleotide alone forms a symmetrically folded hairpin with loop-like properties exhibited by the relatively high emission from a modification in the central 8th repeat and with stem-like properties evident from the relatively low emission intensities from peripheral modifications. Significantly, these hairpin properties are retained in when (CAG)15 is integrated into a duplex. Intrastrand folding by (CAG)15 in the three-way junction contrasts with the open loop adopted by (CAG)8 in the analogous context. This distinction suggests that cooperative interactions in longer repeat tracts overwhelm perturbations to reassert the natural folding propensity. Given that anomalously long repeats are the genetic basis of a large class of inherited neurological diseases, studies with (CAG)-based three-way junctions suggest that their secondary structure is a key factor in the length-dependent manifestation and progression of such diseases.
Keywords: Inherited Neurological Diseases, Trinucleotide Repeats, 2-aminopurine, Fluorescence Spectroscopy, Thermodynamic Measurements
Genetic instability is determined not only by exogenous factors but also by the properties of DNA itself (1, 2). Such inherent mutagenicity was established through association of fragile X syndrome, spinal and bulbar muscular atrophy, and myotonic dystrophy with abnormally long CNG (N = A, T, G, or C) repeats, and subsequently ~ 30 inherited neurological diseases have been linked with expansion of tri-and tetranucleotide repeats beyond critical thresholds (3–6). Linkage between these inherited diseases and sequence length is substantiated by its correlation with phenotypic severity and progression. Furthermore, these mutations are dynamically transmitted, as repeat tracts progressively lengthen through succeeding generations. Beyond the primary information gleaned from DNA sequence and length, secondary structure is the deeper key to this class of genetic diseases, as self-folded conformations are favored by repeated sequences (7). Stem-loop hairpins are favored by CNG repeats and are implicated in repeat expansion via replication, repair, and recombination (5). To illustrate, hairpins that form on single-stranded Okazaki fragments can disrupt coordinated synthesis on the template strands, thereby extending the nascent strand on the leading template through repeated pausing and restarting of DNA polymerase (8). These self-folded moieties can also preferentially recognize and sequester proteins involved in DNA repair, thereby disrupting normal pathways that maintain the DNA integrity (9, 10). Thus, establishing the structures of these alternative forms of DNA is necessary to understand their potential broad-scale biological function.
Within double-stranded DNA, repeated sequences self-associate to produce two distinct structures (11). Slipped forms occur when strands have identical lengths of complementary repeats, and the structures are distinguished from canonical duplex DNA by enhanced sensitivity to nucleases and anomalously fast electrophoretic mobility (12). Their diverse range of structures is stable to challenging environmental changes, thereby suggesting their biological viability. Slipped intermediates form when opposing strands have different lengths, and the resulting three-way junctions are implicated in DNA expansion during replication (13). Within these structures, repeat conformation depends on base sequence, with CTG repeats favoring self-associated hairpins while CAG repeats favor open, solvent-exposed loops, and this difference may originate in the lower thermodynamic stability of CAG vs CTG repeats (7). Our studies show that tract length also dictates conformation.
Toward this goal, the adenine isomer 2-aminopurine is used to develop structural and energetic models of repeated CAG sequences, which are prevalent in many inherited neurological diseases (14). Fluorescent nucleobase analogs are powerful tools for assessing DNA structure and function, and 2-aminopurine is widely used for these purposes (15, 16). Single substitutions with this fluorescent adenine analog do not alter global conformations of CAG based structures, as demonstrated by electrophoresis, spectroscopic, and energetic studies. Structural motifs are identified from their characteristic fluorescence intensities and thermal stability by utilizing the strong effect of base stacking on the fluorescence quantum yield of 2-aminopurine (17, 18). The studies described in this paper are motivated by earlier studies of (CAG)8 (19, 20). The isolated oligonucleotide behaves as the expected stem-loop hairpin, as supported from intensities in the stem region that are comparable to a duplex DNA analog, and intensities in the central loop are comparable to or exceed those of a single-stranded DNA analog. However, when this same sequence is integrated into duplex DNA, the hairpin structure is lost to yield a repeat loop that is highly and uniformly solvent-exposed.
Motivated by how DNA context determines conformation in repeated DNA, we sought to understand how tract length influences secondary structure in slipped intermediate forms of DNA. Utilizing dual structural and energetic perspectives offered by 2-aminopurine substitutions, the secondary structure adopted by (CAG)15 in a three-way junction was determined. Our major finding is that this longer 15 repeat sequence retains the inherent hairpin structure of the isolated oligonucleotide when it is incorporated into the three-way junction. This behavior sharply contrasts with the open loop adopted by the analogous (CAG)8 structure, and such length dependence suggests that repeat length dictates secondary structure and potential biological functions of long repeat tracts.
Materials and Methods
Buffer components (Sigma-Aldrich, St. Louis, MO) and acrylamide (Acros Organics, Belgium) were used as received. The buffer consisted of 10 mM HPO42−/H2PO4− and 50 mM NaCl at pH 7. Oligonucleotides (Integrated DNA Technologies, Coralville, IA) were purified by denaturing 8% PAGE with 7 M urea, TBE 1X buffer at 35 °C, with subsequent visualizing of the samples by UV light on a TLC plate. Desired bands were removed from the gel, electroeluted in TBE buffer, and passed through a NAP-10 desalting column (GE Healthcare, Piscataway, NJ). Samples were then lyophilized and resuspended in water. Oligonucleotide concentrations were determined by extrapolating absorbances at 260 nm of the high-temperature post-transition baselines back to 25 °C. Extinction coefficients of single-stranded oligonucleotides were derived from the nearest neighbor approximation (Table 1). For modified oligonucleotides, extinction coefficients were evaluated as the sum of the extinction coefficient without 2-aminopurine plus the relatively small contribution from free 2-aminopurine (1000 M−1cm−1 at 260 nm) (21). For oligonucleotide and buffer solutions, absorbance was collected at 260 nm as a function temperature on a Cary 300 spectrometer equipped with a multicell holder (Varian, Palo Alto, CA).
Table 1.
Single-Stranded Oligonucleotides.
| Sequence | Lengtha | εb |
|---|---|---|
| 5’-CACCATGCCGGTA TTTAAA CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG TACGTA CTGCAGCTCGAGG-3’ | 95 | 927,000 |
| 5’-CACCATGCCGGTA TTTAAA CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG TACGTA CTGCAGCTCGAGG-3’ c | 95 | 915,200 |
| 5’-CCTCGAGCTGCAG TACGTA CTGCTG – CTGCTG TTTAAA TACCGGCATGGTG-3’ d | 50 | 462,500 |
| 5’-CACCATGCCGGTA TTTAAA CAGCAG – CAGCAG TACGTA CT GCAGCTCGAGG-3’ | 50 | 484,500 |
| 5’-CACCATGCCGGTA TTTAAA CAGCAG – CAGCAG TACGTA CT GCAGCTCGAGG-3’ | 50 | 472,700 |
| 5’-CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG | 45 | 443,800 |
| 5’-CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG CAG e | 45 | 432,000 |
| 5’-G AAA CAG CAG TTTT CTG CTG TTT C-3’ (DS-CAG) | 24 | 210,900 |
| 5’-AAA CAG CAG-3’ (SSβ-CAG) | 9 | 86,300 |
| 5’-CAG CAG CAG-3’ (SS-CAG) | 9 | 78,000 |
Length in bases.
Extinction coefficients (M−1 cm−1) of the unfolded single strands.
Ten variants of this oligonucleotide were used, each with a single substitution of 2-aminopurine, indicated by the underlined bases.
Connecting line represents the region corresponding to the (CAG)15.
Six variants of this oligonucleotide were used, each with a single substitution of 2-aminopurine, indicated by the underlined bases.
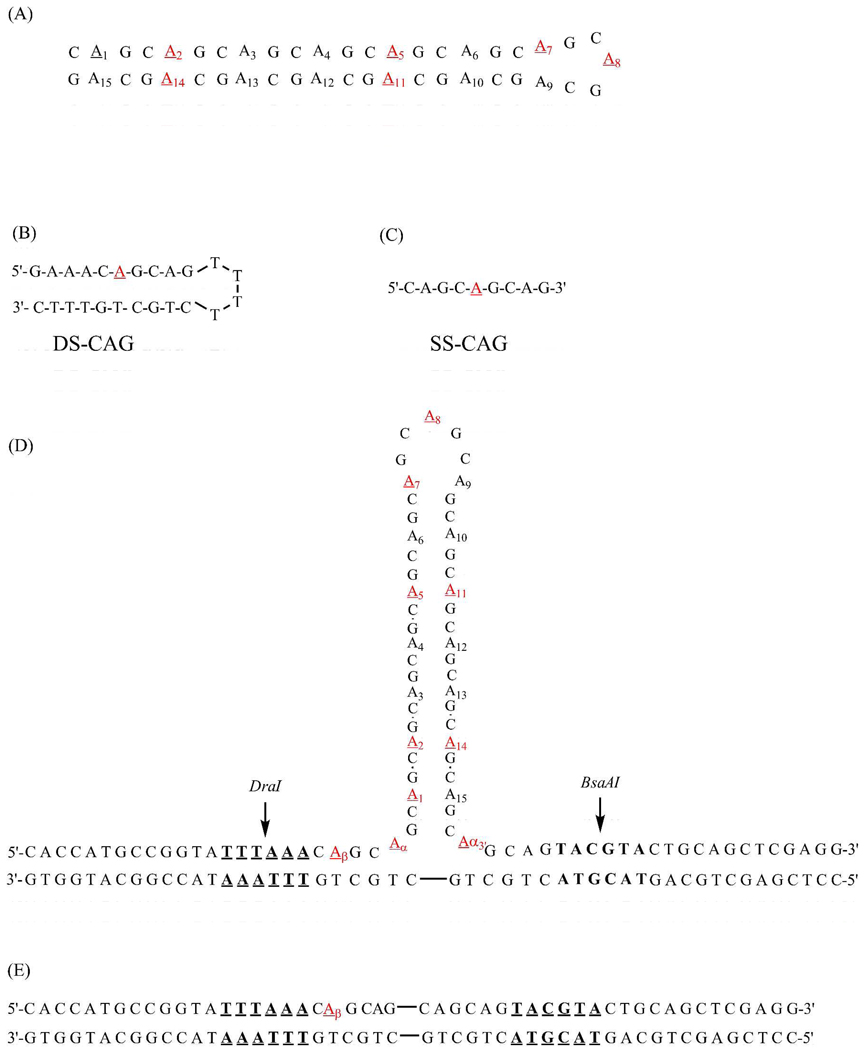
Nomenclature for modified oligonucleotides identifies the position of 2-aminopurine within the primary sequence (Fig. 1). Starting from the 5’terminus, individual replacements of adenine with 2-aminopurine in the isolated (CAG)15 hairpin were made in the 2nd, 5th, 7th, 8th, 11th and 14th repeats, as designated by 2-APHP, 5-APHP, 7-APHP, 8-APHP, 11-APHP, and 14-APHP, respectively. For the three-way junction, substitutions notated as 1-APJ, 2-APJ, 5-APJ, 7-APJ, 8-APJ, 11-APJ, and 14-APJ were made in the 1st, 2nd, 5th, 7th, 8th, 11th, and 14th repeats of the central (CAG)15, respectively. Modifications labeled α–APJ, β-APJ, and α3’-APJ were also made in the two (CAG) repeats in the duplex region that precedes the junction. Three-way junctions were formed by annealing a 95-base oligonucleotide with a central (CAG)15 sequence and flanking 25-base regions that are complementary to a 50-base oligonucleotide (Fig. 1A). These two oligonucleotides conserved the duplex arms used for the three way junction (CAG)8 studies (20). Annealing was performed by heating equimolar amounts of single strands together to 95 °C for 5 minutes with slow cooling to room temperature over a period of more than 12 hours. Purity was verified by 12% polyacrylmide nondenaturing gel electrophoresis. Formation of annealed three-way junctions was monitored with restriction endonucleases BsaAI and DraI (New England BioLabs, Ipswich, MA) using distinct recognition sites on both sides of the repeat tract. At 0.5 µM concentrations in the appropriate buffer (NEB2 from New England BioLabs), DNA samples were mixed with 10–15 units of an enzyme to the total volume of 20 µL and incubated at 37 °C for at least 3 hours. Samples were mixed with SYBR Gold and the gel loading dye and analyzed on the native 12% polyacrylamide gel electrophoresis in TBE buffer. Release of 16 base pair fragment confirmed properly annealed strands (Fig. S1). Similar thermodynamic properties for the modified and unmodified oligonucleotides support minimal perturbation with the single substitutions with 2-aminopurine (Table S1).
Figure 1.
Structures for the hairpin formed by (CAG)15 (A) along with the double-stranded (DS-CAG) (B) and the single-stranded (SS-CAG) (C) structural references. The three-way junction formed with (CAG)15 depicts the DraI and BsaAI restriction enzyme sites in bold and underlined (D). The duplex that comprises the three-way junction includes connecting lines to represent the position of the abstracted repeat sequence (E). For the isolated and integrated forms of (CAG)15, positions of 2-aminopurine substitution are underlined and enumerated.
Fluorescence spectra were collected on a Fluoromax-2 spectrometer (Jobin-Yvon Horiba, Edison, NJ) equipped with DataMax 3.4 software and Neslab RTE-7 circulating bath. Emission was collected at 370 nm using excitation at 307 nm, and the temperature was varied by 0.3 °C with 1 minute equilibration at each temperature (Fig. S2). Thermal denaturation curves accounted for temperature dependent changes in the emission of 2-aminopurine using the single-stranded control SS-CAG (Fig. 1C) (18, 19). Absorbance melting curves were collected at 0.1 °C/min rate using 1 µM concentrations of (CAG)15 and 0.6 µM concentrations of the three-way junctions (Fig. S3). For denaturation profiles exhibiting monophasic profiles, standard van’t Hoff analysis was used to determine the enthalpy and entropy changes. For biphasic transitions, derivative profiles were deconvoluted into gaussian peaks, from which the thermodynamic information was extracted (22). For denaturation of the ith domain, the enthalpy change (ΔHi) at the melting temperature (Tm) is:
where R is the gas constant, hi is the peak amplitude, and ΔAi is the peak area. Entropy changes accounted for the concentration dependence using
where c0 is the initial strand concentration (23). Average thermodynamic parameters and standard deviations were derived from a minimum of three thermal denaturation profiles that were collected using separate samples.
For acrylamide studies, standard Stern-Volmer analysis was an adequate model to determine the static quenching constants (24). Concentrations up to 0.1 M acrylamide were used. Oligonucleotide concentrations were 0.3–0.5 µM, and average quenching constants with standard deviations were calculated from at least three replicate experiments.
Results
(CAG)15 Hairpin
Six variants with 2-aminopurine establish the propensity of (CAG)15 to fold into a stem-loop hairpin (7). A minimal degree of structural perturbation by this adenine isomer is supported by similar circular dichroism spectra of modified and unmodified oligonucleotides (Fig. S4). The spectra are characteristic of B-form DNA, suggesting a duplex stem that is stabilized by the high proportion of G/C pairs (25). Base stacking in the folded structure is also indicated by hyperchromic absorbance changes at high temperatures. Derived thermodynamic parameters do not vary with 2-aminopurine modification, with the unmodified hairpin having ΔH = 38.7 ± 1.8 kcal/mole and ΔS = 117 ± 5 cal/(mole K) at 58.0 ± 0.3 °C and the six variants having an average ΔH = 37.9 ± 1.6 kcal/mole and ΔS = 114 ± 5 cal/(mole K) at 58.2 ± 0.8 °C (Table 2).
Table 2.
Thermodynamic Data for Thermal Denaturation and Acrylamide Quenching Constants for (CAG)15 Sequencesa
| Positionb | Tm,abs (°C) |
ΔHabs (kcal/mol) |
ΔSabs (cal/mol K) |
Tm,fluor (°C) |
ΔHfluor (kcal/mol) |
ΔSfluor (cal/mol K) |
Kq (M−1)f |
|---|---|---|---|---|---|---|---|
| 2-APHP | 58.4±0.3 | 35.9±0.9 | 108±3 | 57.7±0.3 | 38.2±1.8 | 116± 5 | 3.7±0.4 |
| 5-APHP | 58.9±0.6 | 38.1±2.9 | 114±9 | 58.1±0.3 | 41.8±1.6 | 127± 5 | 5.8±0.6 |
| 7-APHP | 57.4±0.8 | 39.2±0.7 | 118±2 | 58.7±1.0 | 41.2±4.6 | 124± 14 | 6.8±0.6 |
| 8-APHP | 57.8±0.4 | 39.0±0.6 | 118±2 | NDe | ND | ND | 8.0±0.5 |
| 11-APHP | 59.4±1.0 | 35.9±0.9 | 108±3 | 58.4±0.8 | 39.2±2.3 | 119± 7 | 5.5±0.4 |
| 14-APHP | 57.4±0.2 | 39.4±0.4 | 119±1 | 57.5±0.8 | 42.7±1.4 | 129± 4 | 4.5±0.7 |
| Averagec | 58.2±0.8 | 37.9±1.6 | 114±5 | 58.0±0.5 | 40.6±1.9 | 123±5 | |
| (CAG)15d | 58.0±0.3 | 38.7±1.8 | 117±5 |
Subscripts “abs” and “fluor” indicate measurements using absorbance at 260 nm and fluorescence using λex = 307 nm/λem = 370 nm, respectively. Standard deviations are derived from a minimum of three measurements. Melting temperatures (Tm), enthalpy changes (ΔH), and entropy changes (ΔS) were derived from absorbance and fluorescence changes. Quenching constants (Kq) were derived from acrylamide quenching of 2-aminopurine fluorescence.
See Figure 1A for location of modifications in (CAG)15.
Average data for the absorbance and fluorescence measurements for the above 6 variants.
Thermodynamic data derived for the unmodified (CAG)15 oligonucleotide.
Values not determined because of poorly defined denaturation profile.
Quenching constants for the double-stranded (DS-CAG) and single-stranded standards (SS-CAG) are is 4.2 ± 1.7 M−1 and 9.6 ± 1.0 M−1, respectively.
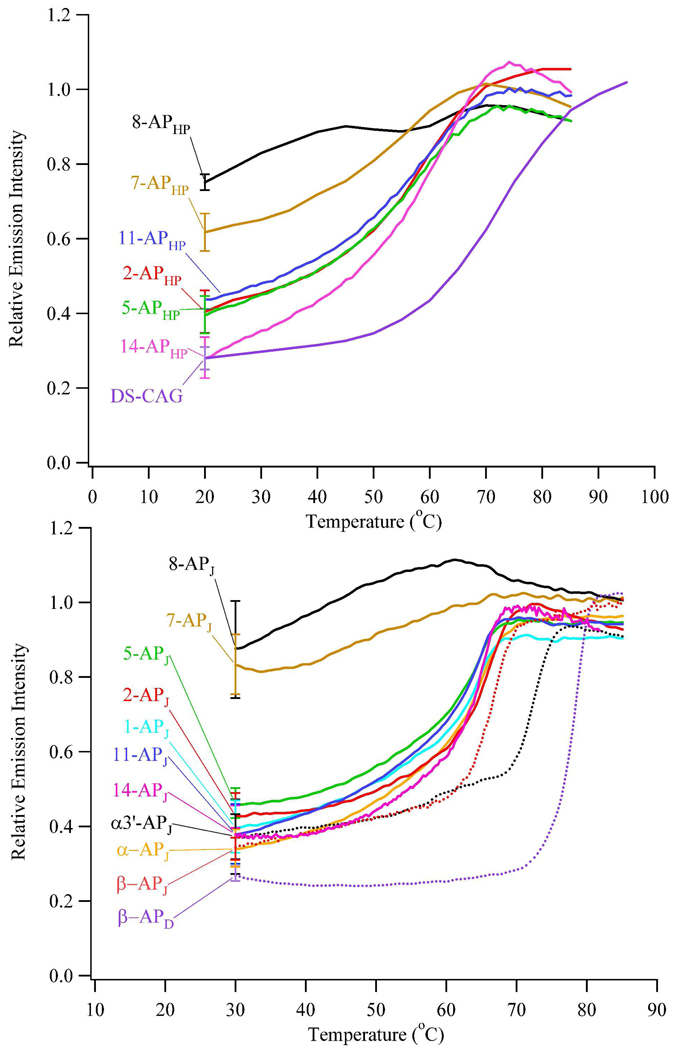
The secondary structure of (CAG)15 was established through the solvent accessibility of individually substituted 2-aminopurines, and two types of references relate emission intensities with particular structural moieties. First, a hairpin oligonucleotide with a 2-aminopurine/thymine pair in the stem defines a lower limit of solvent exposure (DS-CAG, Fig. 1B). In this arrangement, fluorescence is quenched through base stacking, although stacking is less efficient when compared with the corresponding adenine/thymine pair (26). Second, a short 9-base (CAG)3 oligonucleotide with a centrally placed 2-aminopurine defines an upper limit of solvent exposure (SS-CAG, Fig. 1C). This length precludes folding and base stacking, thereby enhancing the fluorescence quantum yield of 2-aminopurine. In addition, this sequence reflects the environment in the repeat tracts, in which repeats with 2-aminopurine are flanked by CAG trinucleotides. This strand exhibits no tendency to self-associate based on fluorescence studies conducted at higher and lower concentrations and based on similar temperature dependent fluorescence changes for this short oligonucleotide and free 2-aminopurine (19). Fluorescence intensities follow the order: DS-CAG < 14-APHP ~ 2-APHP ~ 5-APHP ~ 11-APHP < 7-APHP < 8-APHP < SS-CAG, where the extreme intensities are associated with the double-stranded and single-stranded references, respectively. This trend advances a hairpin structure (Fig. 2 and S5). First, substitutions in the 2nd, 5th, 11th, and 14th repeats that flank the center exhibit intensities that are more similar to the double-stranded reference. This similarity suggests that base stacking is a significant driving force for folding, consistent with characteristic B-form signature in circular dichroism spectra (Fig. S4). Higher intensities for these emissive variants relative to the double-stranded reference suggest that purine-purine mismatches increase solvent exposure (27). Second, substitutions in the 7th and 8th repeats show that intensities increase toward the center of the primary sequence and approach the reference intensity of single-stranded DNA. Together, these observations are consistent with symmetric folding to produce duplex stem connected by a central loop.
Figure 2.
Emission intensities of the (CAG)15 hairpin (top) and its three-way junctions (bottom) with 2-aminopurine as a function of temperature using λex = 307 nm and λem = 370 nm. These intensities are corrected for the inherent temperature dependent changes of 2-aminopurine using the single-stranded DNA reference (SS-CAG, Fig. 1C).
All 2-aminopurine oligonucleotides attain a common denatured state at sufficiently high temperature, and temperature dependent fluorescence provides energetic signatures of the stem and loop. An inherent temperature effect on fluorescence quantum yield is removed by also collecting emission from a single-stranded DNA reference with limited base stacking (Fig. 1C) (18). Two types of environments are revealed in fluorescence thermograms of (CAG)15: a relatively unstructured loop evidenced by modest intensity changes for 8-APHP and a base stacked stem as indicated by enhanced fluorescence for 2-APHP, 5-APHP, 11-APHP, and 14-APHP. Furthermore, a transition between these motifs is evident from intermediate enhancement of 7-APHP (Fig. 2A). Thermodynamic analysis used limiting intensities to derive the fractional conversion from folded to unfolded forms, and resulting enthalpy and entropy changes show no distinction between the hyperfluorescent oligonucleotides with an average ΔH = 40.6 ± 1.9 kcal/mole and ΔS = 123 ± 5 cal/(mole K) at 58.1 ± 0.5 °C (Table 2). This similarity indicates cooperative denaturation, as also corroborated by similar globally-based thermodynamic parameters determined from absorbance measurements (Table 2). Derived van’t Hoff parameters agree with previously reported values, although they differ from model-free parameters determined by calorimetry (25, 28). Cooperative unfolding deduced from our experiments suggests that other factors such as heat capacity changes due to solvation effects may be important in the unfolding process (28).
To extrinsically probe the hairpin structure of (CAG)15, 2-aminopurine fluorescence was quenched with acrylamide (Table 2). Solvent accessibility dictates the degree of quenching, and the single- and double-stranded references again distinguish the structural motifs in (CAG)15. Substitutions in the 2nd and 14th repeats are more protected and quenched least efficiently because they behave most similarly to the double-stranded reference with a quenching constant of 4.2 ± 1.7 M−1. Converging toward the center, quenching efficiency increases and culminates for 8-APHP, which behaves most similarly to the single-stranded reference with a quenching constant of 9.6 ± 1.0 M−1. This behavior suggests a gradual opening of the duplex stem in the vicinity of the loop, as also observed for (CAG)8 (19).
(CAG)15 Three-Way Junction
By establishing the inherent folding propensity of (CAG)15 alone, its degree of perturbation within the three-way junction is evaluated. Using identical substitutions made in the isolated form of (CAG)15, fluorescence studies distinguish two environments in the integrated form (Fig. 1D and 2). Stem formation is demonstrated by intensities from 2-APJ/14-APJ and 5-APJ/11-APJ that are most similar to a double-stranded DNA reference. Intrastrand folding is further substantiated by similar fluorescence intensities from these distantly placed modifications and by their enhanced fluorescence that accompanies denaturation (Figs 2 and S6). Loop formation in the repeat tract is supported by two measurements on 8-APJ: its emission intensity changes little with temperature and is most similar to the single-stranded DNA reference. Transition between these two motifs is corroborated by an intermediate fluorescence intensity and a weak, monophasic response to temperature that is exhibited by 7-APJ. Importantly, these same intensities and intensity changes are exhibited by the analogous variants of the (CAG)15 oligonucleotide alone, indicating that the inherent structure of this repeated sequence is retained when it is incorporated in the duplex. In addition to probes in the repeat region of the three-way junction, the structure and stability in the supporting duplex arms were evaluated using substitutions in the β, α, and α3’ repeats. All three modifications emit similarly to the double-stranded DNA reference. Thus, base stacking is a dominant interaction in the supporting arms, and this effect extends into the repeat region, as indicated by comparably low intensities from 1-APJ. These intensities are slightly higher than the double-stranded reference and increase in the vicinity of the repeat tract, which indicates that the repeat tract influences base pairing and stacking (Fig. 2).
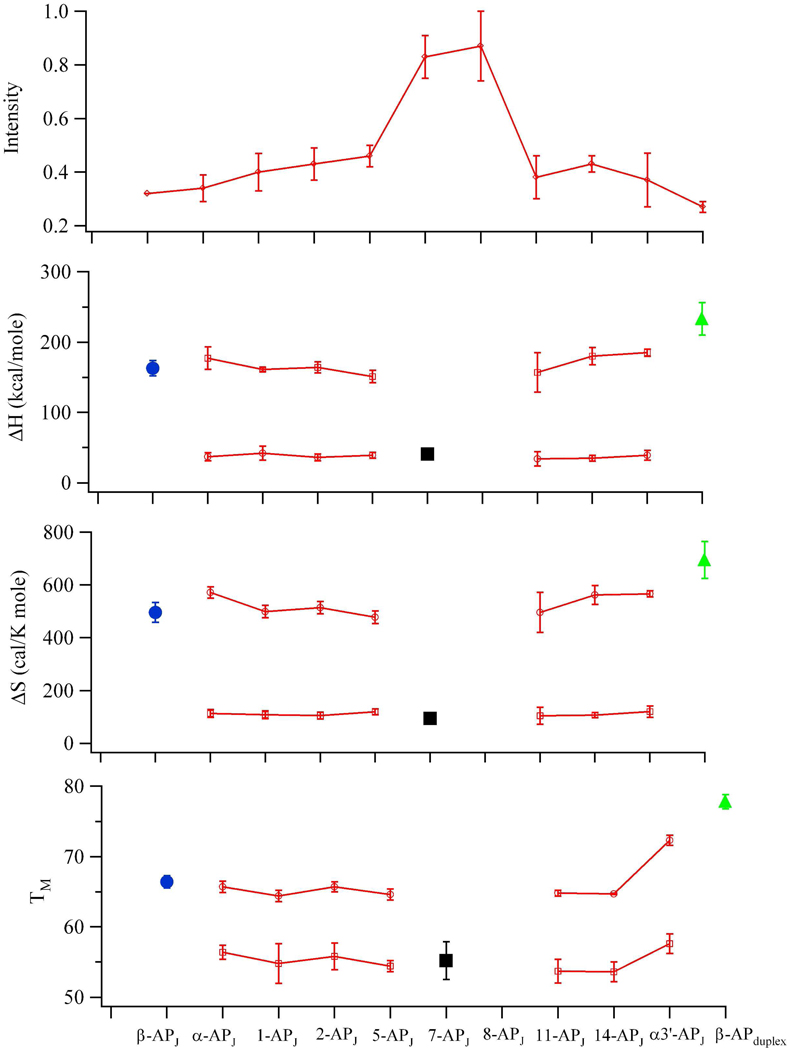
Segregation into distinct domains is also supported through stability measurements. Absorbance studies at 260 nm exhibit two resolved transitions during thermal denaturation (Fig. S3 and Table S1). The high temperature transition at 71.9 ± 0.6 °C has ΔH = 235 ± 3 kcal/mole and ΔS = 712 ± 8 cal/(mole K), which is similar to the ΔH = 252 ± 12 kcal/mole and ΔS = 694 ± 30 cal/(mole K) measured at 75.4 ± 1.1 °C for the component duplex without (CAG)15 (Fig. 1E and Table S1). Their differences may reflect compromised base stacking and pairing at the junction. In relation to (CAG)15 alone, the low temperature transition at 65.6 ± 0.7°C is more thermodynamically restrained with ΔH = 110 ± 9 kcal/mole and ΔS = 326 ± 24 cal/(mole K), and this difference may be due to anchoring and overall stabilization by the duplex arms. To further clarify these structural motifs and the transitional regions between them, thermodynamic analysis was conducted using fluorescence (Fig. 3 and Table 3). As in the absorbance measurements, the majority of fluorescence thermograms exhibit biphasic profiles (Fig. S2). Within these profiles, melting temperatures, enthalpy changes, and entropy changes for the two transitions are similar across the different modifications, indicating cooperative relaxation within the domains. The breadth of the lower temperature transition may be related to local premelting of the mismatched pairs prior to overall denaturation of the hairpin region (19). The structural basis of these two transitions is established via two peripheral modifications with monophasic profiles (Fig. 3). First, β-APJ within the duplex arms is distant from the junction, and its single transition exhibits thermodynamic properties that mimic the high temperature transition. This structural and thermodynamic correlation suggests that the high temperature transition reflects denaturation of the duplex arms. Duplex integrity is altered by the repeat sequence, as shown by a higher fluorescence intensity and a lower thermodynamic stability for β-APJ in relation to its duplex analog without (CAG)15 (Figs. 1D, 1E, and 3 and Table 3). The second peripheral reference is provided by 7-APJ, which is close to apex of the hairpin and thus also distant from the junction. Its monophasic transition reflects the thermodynamic properties associated with the low temperature transitions in the other variants, thus the low temperature transition is assigned to denaturation of the repeat stem. This assignment is validated by comparable thermodynamic properties for the isolated (CAG)15 hairpin. For substitutions in the 2nd, 5th, 11th, and 14th repeats, the three-way junction has an average ΔH = 36.0 ± 2.1 kcal/mole and an average ΔS = 109 ± 7 cal/(mole K) at 56 ± 2 °C, which are comparable to the analogous averages for (CAG)15 alone of ΔH = 41 ± 2 kcal/mole and an average ΔS = 123 ± 6 cal/(mole K) at 57.9 ± 0.4 °C. Further reflecting the properties of the hairpin, thermodynamic integrity is almost lost by the peripheral 8th repeat, as indicated by its weak and structureless response to temperature changes. These similarities between the integrated and isolated forms of (CAG)15 provide strong evidence that the inherent folding propensity of this longer repeat tract is retained in the three-way junction.
Figure 3.
Summary of fluorescence intensities and thermodynamic results for the (CAG)15 three-way junction. (Top panel) Emission intensities increase in the center of the primary sequence, indicative of stem-loop formation. (Bottom three panels) Enthalpy (ΔH) and entropy (ΔS) changes and melting temperatures (Tm) measured throughout the three-way junction. Substitutions in the α, 1st, 2nd, 5th, 11th, 14th, and α3’ repeats (open red squares) exhibit similar thermodynamic changes. Modifications in the β (blue circles) and 7th (black squares) repeats exhibit monophasic transitions that mimic the high and low temperature transitions, respectively, in the other fluorescent variants. Parameters associated with double-stranded DNA without (CAG)15 are indicated by green triangles. Error bars represent standard deviations derived from at least three measurements.
Table 3.
Thermodynamic Data for Thermal Denaturation and Acrylamide Quenching Constants for (CAG)15-Based Three-Way Junctionsa
| positionb | Tm1 (°C) | ΔH1 (kcal/mol) |
ΔS1 (cal/mol/K) |
Tm2 (°C) |
ΔH2 (kcal/mol) |
ΔS2 (cal/mol/K) |
Kq (M−1)d |
|---|---|---|---|---|---|---|---|
| β-APJ | 66.4±0.9 | 163±11 | 496±38 | 4.6±0.3 | |||
| α-APJ | 56.4±1.0 | 37±6 | 113±15 | 65.7±0.8 | 177±16 | 572±21 | 3.9±0.2 |
| 1-APJ | 54.8±2.8 | 42±10 | 108±15 | 64.4±0.8 | 161±3 | 499±24 | 5.3±0.3 |
| 2-APJ | 55.8±1.9 | 36±5 | 105±13 | 65.7±0.7 | 164±8 | 514±24 | 4.1±0.2 |
| 5-APJ | 54.4±0.8 | 39±4 | 119±11 | 64.6±0.8 | 151±2 | 478±4 | 5.6±0.2 |
| 7-APJ | 55.2±2.7 | 41±4 | 94±9 | 8.7±0.2 | |||
| 8-APJ | NDc | ND | ND | ND | ND | ND | 8.3±0.2 |
| 11-APJ | 53.7±1.7 | 34±10 | 104±32 | 64.8±0.4 | 157±28 | 496±76 | 4.6±0.2 |
| 14-APJ | 53.6±1.4 | 35±4 | 107±1 | 64.7±0.1 | 180±12 | 562±36 | 4.0±0.2 |
| α3’-APJ | 57.6±1.4 | 39±7 | 120±21 | 72.3±0.7 | 185±4 | 565±10 | 4.6±0.2 |
| β-APD | 77.8±1.0 | 233±23 | 695±70 |
Parameters measured using fluorescence at λex = 307 nm/λem = 370 nm. Standard deviations are derived from a minimum of three measurements. Melting temperatures (Tm), enthalpy changes (ΔH), and entropy changes (ΔS) were derived from fluorescence changes. Quenching constants (Kq) were derived from acrylamide quenching of 2-aminopurine fluorescence. Subscripts 1 and 2 correspond to the low and high temperature transitions, respectively.
See Figure 1D for location of modifications in (CAG)15.
Values not determined because of poorly defined denaturation profile.
Quenching constants for the double-stranded (DS-CAG) and single-stranded standards (SS-CAG) are 4.2 ± 1.7 M−1 and 9.6 ± 1.0 M−1,respectively.
Acrylamide quenching constants also support a repeat hairpin that emanates from the duplex arms (Table 3). Modifications in the 7th and 8th positions have highest quenching constants that are similar to the single-stranded reference, thus consistent with higher base exposure in a loop. Quenching constants of 1-APJ, 2-APJ, 5-APJ, 11-APJ, and 14-APJ, α–APJ, β-APJ, and α’-APJ are similar to the fully sequestered base in the double-stranded reference. These features were also exhibited by (CAG)15 alone.
Discussion
These studies demonstrate the inherent propensity of longer CAG repeats to fold, both as isolated oligonucleotides and within three-way junctions. From a structural standpoint, fluorescence intensity and acrylamide quenching studies of six 2-aminopurine variants support hairpin formation in both contexts. Solvent-sequestered, stem-like properties are exhibited by substitutions in the 2nd/14th and 5th/11th repeats, which have comparably low intensities and quenching constants in relation to double-stranded DNA analogs. Adjoining loops encompassing the 7th and 8th repeats are discerned by their high level of solvent exposure when compared with the single-stranded DNA reference. Hairpin folding in both forms of (CAG)15 is further substantiated through stability studies. Most importantly, similar thermodynamic properties for the isolated hairpin and the repeat tract in the three-way junction support the formation of a repeat hairpin domain within the three-way junction. The global structure of the three-way junction is inferred by mapping solvent exposure through the junction region. Base stacking and pairing is maintained throughout the junction, and prior studies of DNA three-way junctions suggest that the three arms should be fully extended (29). Thus, for the (CAG)15 based three-way junction, two arms are canonical duplexes while the third is the repeat hairpin (Fig. 1C).
These results broaden an understanding of repeat folding in slipped intermediates that are postulated to develop during DNA replication. CAG tracts incorporated into duplex DNA adopt associated hairpin and unassociated loop forms (13). In support of unstructured loops, the repeat region is preferentially digested by single-strand specific enzymes (13). In vivo studies using over-expressed proteins and in vitro studies that measure DNA structure via electron microscopy support a preferential affinity of single-strand binding proteins for CAG repeats (13, 30). Spectroscopic based studies using 2-aminopurine showed that (CAG)8 within a three-way junction is uniformly open and solvent exposed, and the results suggest that the duplex component dominates both structure and stability within the composite three-way junction (18). Collectively, these studies suggest that CAG repeats inherently favor open loops, possibly induced by purine-purine mismatches that are expected to reduce hairpin stability (7). However, such structures should be considered in a larger context, as the longer (CAG)15 retains its native hairpin structure when incorporated in a three-way junction. Cooperative interactions that drive intrastrand association may be the origin of this stark structural difference that depends on the number of repeats. For isolated hairpins, stability increases with the number of repeats because of the more favorable enthalpic interactions from base stacking and pairing (25, 28). Our results suggest that cooperativity impacts secondary structure in repeat tracts of three-way junctions. Assessing the significance of cooperativity on folding could be accomplished using interruptions such as CAA in place of CAG, which are known to influence in vivo genetic instability (31, 32). Because CAG repeats form one of the least stable folded hairpins, cooperative effects are expected to be more significant for other repeated sequences.
Conclusion
At the genetic level, structures adopted by abnormally long repeated DNA sequences are implicated in a large class of inherited neurological diseases, and slipped intermediates that form during DNA replication provide one avenue for sequence expansion. Folding that facilitates expansion depends on base sequence and DNA context, and our studies demonstrate that sequence length also dictates intrastrand association within the repeat tract. We suggest that base stacking promotes base pairing between distant repeats and cooperative interactions drive such folding in longer repeat tracts. Length-dependent folding is expected to influence other repeat tracts, thereby impacting processes that produce and recognize abnormally long repeat tracts.
Supplementary Material
Acknowledgement
We thank the National Science Foundation (CHE-0718588) for primary support of this work. We are grateful to the National Institutes of Health (R15GM071370), National Science Foundation (CBET-0853692), Henry Dreyfus Teacher-Scholar Awards Program, and the National Institutes of Health (P20 RR-016461 from the National Center for Research Resource) for additional support throughout this work. JTP received partial sabbatical support through matching commitments to an NSF RII Cooperative Agreement (EPS-0903795).
Footnotes
Supporting Information Available: Supplemental figures describing structural characterization of the three-way junction, deconvolution of fluorescence and absorbance thermograms, circular dichroism spectra of (CAG)15, and composite fluorescence spectra of (CAG)15 and of the three-way junction variants and a table summarizing thermodynamic information related to the three-way junction are available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bacolla A, Wells RD. Non-B DNA conformations as determinants of mutagenesis and human disease. Mol. Carcinog. 2009;48:273–285. doi: 10.1002/mc.20507. [DOI] [PubMed] [Google Scholar]
- 2.Wang G, Vasquez KM. Non-B DNA structure-induced genetic instability. Mutation Research/Fundamental and Molecular Mechanisms of Mutagenesis. 2006;598:103–119. doi: 10.1016/j.mrfmmm.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 4.Castel AL, Cleary JD, Pearson CE. Repeat instability as the basis for human diseases and as a potential target for therapy. Nat. Rev. Mol. Cell Biol. 2010;11:165–170. doi: 10.1038/nrm2854. [DOI] [PubMed] [Google Scholar]
- 5.Kovtun IV, McMurray CT. Features of trinucleotide repeat instability in vivo. Cell Res. 2008;18:198–213. doi: 10.1038/cr.2008.5. [DOI] [PubMed] [Google Scholar]
- 6.Gatchel JR, Zoghbi HY. Diseases of Unstable Repeat Expansion: Mechanisms and Common Principles. Nat. Rev. Genet. 2005;6:743–755. doi: 10.1038/nrg1691. [DOI] [PubMed] [Google Scholar]
- 7.Mitas M. Trinucleotide repeats associated with human disease. Nucleic Acids Res. 1997;25:2245–2254. doi: 10.1093/nar/25.12.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirkin SM. DNA structures, repeat expansions and human hereditary disorders. Current Opinion in Structural Biology. 2006;16:351–358. doi: 10.1016/j.sbi.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 9.Kovtun IV, McMurray CT. Trinucleotide expansion in haploid germ cells by gap repair. Nat. Genet. 2001;27:407–411. doi: 10.1038/86906. [DOI] [PubMed] [Google Scholar]
- 10.Panigrahi GB, Slean MM, Simard JP, Gileadi O, Pearson CE. Isolated short CTG/CAG DNA slip-outs are repaired efficiently by hMutSβ, but clustered slip-outs are poorly repaired. Proc. Natl. Acad. Sci. U. S. A. 2010;107:12593–12598. doi: 10.1073/pnas.0909087107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sinden RR, Potaman VN, Oussatcheva EA, Pearson CE, Lyubchenko YL, Shlyakhtenko LS. Triplet repeat DNA structures and human genetic disease: dynamic mutations from dynamic DNA. J. Biosci. 2002;27:53–65. doi: 10.1007/BF02703683. [DOI] [PubMed] [Google Scholar]
- 12.Pearson CE, Sinden RR. Alternative structures in duplex DNA formed within the trinucleotide repeats of the myotonic dystrophy and fragile X loci. Biochemistry. 1996;35:5041–5053. doi: 10.1021/bi9601013. [DOI] [PubMed] [Google Scholar]
- 13.Pearson CE, Tam M, Wang Y-H, Montgomery SE, Dar AC, Cleary JD, Nichol K. Slipped-strand DNAs formed by long (CAG)-(CTG) repeats: Slipped-out repeats and slip-out junctions. Nucleic Acids Res. 2002;30:4534–4547. doi: 10.1093/nar/gkf572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson CE, Edamura KN, Cleary JD. Repeat instability: mechanisms of dynamic mutations. Nat. Rev. Genet. 2005;6:729–742. doi: 10.1038/nrg1689. [DOI] [PubMed] [Google Scholar]
- 15.Sinkeldam RW, Greco NJ, Tor Y. Fluorescent Analogs of Biomolecular Building Blocks: Design, Properties, and Applications. Chem. Rev. 2010;110:2579–2619. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rist MJ, Marino JP. Fluorescent Nucleotide Base Analogs as Probes of Nucleic Acid Structure, Dynamics and Interactions. Curr. Org. Chem. 2002;6:775. [Google Scholar]
- 17.Ward DC, Reich E, Stryer L. Fluorescence studies of nucleotides and polynucleotides. I. Formycin, 2-aminopurine riboside, 2,6-diaminopurine riboside, and their derivatives. J. Biol. Chem. 1969;244:1228–1237. [PubMed] [Google Scholar]
- 18.Ballin JD, Bharill S, Fialcowitz-White EJ, Gryczynski I, Gryczynski Z, Wilson GM. Site-Specific Variations in RNA Folding Thermodynamics Visualized by 2-Aminopurine Fluorescence. Biochemistry. 2007;46:13948–13960. doi: 10.1021/bi7011977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Degtyareva NN, Reddish MJ, Sengupta B, Petty JT. Structural Studies of a Trinucleotide Repeat Sequence Using 2-Aminopurine. Biochemistry. 2009;48:2340–2346. doi: 10.1021/bi802225y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Degtyareva NN, Barber CA, Sengupta B, Petty JT. Context dependence of trinucleotide repeat structures. Biochemistry. 2010;49:3024–3030. doi: 10.1021/bi902043u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fox JJ, Wempen I, Hampton A, Doerr IL. Thiation of Nucleosides. I. Synthesis of 2-Amino-6-mercapto-9-β-D-ribofuranosylpurine (“Thioguanosine”) and Related Purine Nucleosides1. J. Am. Chem. Soc. 1958;80:1669–1675. [Google Scholar]
- 22.Yen WS, Blake RD. Analysis of high-resolution melting (thermal dispersion) of DNA. Methods. Biopolymers. 1980;19:681–700. doi: 10.1002/bip.1980.360190316. [DOI] [PubMed] [Google Scholar]
- 23.Mergny J-L, Lacroix L. Analysis of Thermal Melting Curves. Oligonucleotides. 2003;13:515–537. doi: 10.1089/154545703322860825. [DOI] [PubMed] [Google Scholar]
- 24.Lakowicz JR. Principles of Fluorescence Spectroscopy. New York: Plenum Press; 1983. [Google Scholar]
- 25.Paiva AM, Sheardy RD. Influence of sequence context and length on the structure and stability of triplet repeat DNA oligomers. Biochemistry. 2004;43:14218–14227. doi: 10.1021/bi0494368. [DOI] [PubMed] [Google Scholar]
- 26.Lycksell PO, Graslund A, Claesens F, McLaughlin LW, Larsson U, Rigler R. Base pair opening dynamics of a 2-aminopurine substituted Eco RI restriction sequence and its unsubstituted counterpart in oligonucleotides. Nucleic Acids Res. 1987;15:9011–9025. doi: 10.1093/nar/15.21.9011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Arnold FH, Wolk S, Cruz P, Tinoco I., Jr Structure, dynamics, and thermodynamics of mismatched DNA oligonucleotide duplexes d(CCCAGGG)2 and d(CCCTGGG)2. Biochemistry. 1987;26:4068–4075. doi: 10.1021/bi00387a049. [DOI] [PubMed] [Google Scholar]
- 28.Amrane S, Sacca B, Mills M, Chauhan M, Klump HH, Mergny J-L. Length-dependent energetics of (CTG)n and (CAG)n trinucleotide repeats. Nucleic Acids Res. 2005;33:4065–4077. doi: 10.1093/nar/gki716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lilley DMJ. Analysis of branched nucleic acid structure using comparative gel electrophoresis. Q. Rev. Biophys. 2008;41:1–39. doi: 10.1017/S0033583508004678. [DOI] [PubMed] [Google Scholar]
- 30.Andreoni F, Darmon E, Poon WCK, Leach DRF. Overexpression of the single-stranded DNA-binding protein (SSB) stabilises CAG-CTG triplet repeats in an orientation dependent manner. FEBS Letters. 2010;584:153–158. doi: 10.1016/j.febslet.2009.11.042. [DOI] [PubMed] [Google Scholar]
- 31.Jarem DA, Huckaby LV, Delaney S. AGG Interruptions in (CGG)n DNA Repeat Tracts Modulate the Structure and Thermodynamics of Non-B Conformations in Vitro. Biochemistry. 2010;49:6826–6837. doi: 10.1021/bi1007782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sobczak K, Krzyzosiak WJ. CAG Repeats Containing CAA Interruptions Form Branched Hairpin Structures in Spinocerebellar Ataxia Type 2 Transcripts. J. Biol. Chem. 2005;280:3898–3910. doi: 10.1074/jbc.M409984200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.