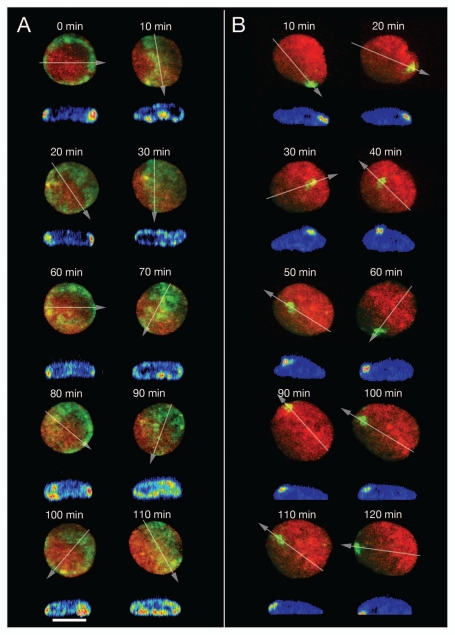
Figure 3.
Major changes of CT proximity patterns in RPE-1 interphase nuclei during complex large-scale rotational movements of chromatin assemblies. (A) A ring of paGFP fluorescent chromatin was induced at the 2D rim of a flat-ellipsoidal RPE-1 cell nucleus (time 0). 3D images were taken every 10 min for a total period of 110 min observation time. Note the drastic, repetitive changes of the fluorescent patterns noted in the 2D projections of this image sequence from ring-shaped to broad bands etc., indicating repeated rotations along an perpendicular axis. Since the flat-ellipsoidal shape of the nucleus was maintained during these rotations of entire CT assemblies, proximity patterns changed accordingly (compare Fig. 7D). Virtual cross-sections performed through 3D nuclear reconstructions along the lines indicated by arrows demonstrate that the mass of fluorescent chromatin remained at the nuclear envelope during these rotations indicating that radial interphase chromatin arrangements were maiantained during complex nuclear rotation. (B) Photoactivation of paGFP in a small area at the nuclear rim as a marker for major chromatin movements at the nuclear periphery. At face value 2D projections of x, y-nuclear images recorded at different times after photoactivation indicate movements of marker chromatin away from the nuclear rim (40–60 min) and back to it (70–130 min); virtual x, z-cross-sections through 3D nuclear reconstructions along lines indicated in the 2D projections by arrows demonstrate that the marker chromatin always stayed at the nuclear periphery. During the whole observation period the nucleus performed a counter-clockwise rotation of about 630° around an axis perpendicular to the growth surface and simultaneously another rotation around an axis parallel to it.