Abstract
Many autoimmune diseases share a genetic association with the presence or absence of HLA-DQB1*0602, including type I diabetes, multiple sclerosis, and narcolepsy. High resolution HLA typing to determine the presence of this allele is cumbersome and expensive by currently available techniques. We present a real-time PCR assay for the identification of HLA-DQB1*0602, using sequence-specific primers and probes, that provides rapid and sensitive identification of this allele, involves minimal hands-on time, and provides a major cost savings compared to existing methods. The assay allows the simultaneous determination of both the presence and the number of copies of this allele. Since there is no post-PCR handling, the risk of contamination is avoided. We have validated the assay using 44 blinded and 32 unblinded samples, previously typed by standard techniques, which were identified with 100% accuracy, sensitivity, and specificity. Further, using a narcolepsy cohort of 734 subjects, we demonstrated the robustness of the assay to analyze DNA isolated from buccal swabs, demonstrating the applicability of this assay as an alternative approach to traditional HLA typing methods.
Keywords: Genotyping, HLA-DQB1*0602, HLA typing, Multiple Sclerosis, Narcolepsy, QPCR-SSPP, Real-time PCR, Systemic Lupus Erythematosus, TaqMan, Type 1 Diabetes Mellitus
Introduction
The human histocompatibility locus antigen (HLA) plays a key role in autoimmune disease etiology. As one component of the trimolecular complex (MHC-peptide-T cell receptor), the presence of specific HLA alleles determines the repertoire of peptide epitopes that can be presented, restricting the specificity of reactive T cells (1–4). HLA-DQB1*0602 is associated with various autoimmune and other diseases, including narcolepsy (1;5–7), multiple sclerosis (8;9), systemic lupus erythematosus (10–12), sarcoidosis (13), primary sclerosing cholangitis (14), and gastric, cervical, and intraepithelial cancers (15–18). HLA-DQB1*0602 is also strongly associated with dominant protection against type I diabetes (19).
Traditional molecular methods for HLA typing, such as polymerase chain reaction (PCR) with sequence-specific oligonucleotide probes (PCR-SSO) (20), require an initial PCR amplification step, followed by post-PCR hybridization using specific probes. The resultant typing is generally low resolution. PCR with sequence-specific primers (PCR-SSP) (21) and sequence-based typing (PCR-SBT) are both able to resolve HLA alleles with high resolution (22), but both are labor intensive and time consuming. Further, each of these methods requires post-PCR handling which introduces the potential for cross-contamination of samples and reagents.
We previously introduced a real-time PCR assay for rapid HLA-DR4 subtyping (23). The technique, called QPCR-SSPP (quantitative PCR with sequence-specific primers and probes), is performed in a sealed PCR plate and does not require any post-PCR manipulation. In this report we adapt the use of QPCR-SSPP to the rapid typing of HLA-DQB1*0602, with amplification and detection steps carried out simultaneously in real time. The assay involves minimal handling steps, is less expensive than other methods, and is rapid, sensitive, and accurate.
Materials and Methods
Isolation of DNA
Peripheral blood or buccal swabs were obtained from healthy adults and autoimmune disease patients after obtaining informed consent. Genomic DNA was isolated using the QIAamp® DNA Blood Mini Kit (Qiagen, Inc.).
Primers and Probes
Primers and probes (shown in Table 1) were designed within exon 2 of the HLA-DQB1 locus, using Primer Express™ v2.0 (Applied Biosystems, Inc.), based on the sequence alignments of Release 2.21.0 of the IMGT/HLA Sequence Database (updated April 8, 2008; see http://www.ebi.ac.uk/imgt/hla/align.html) (24). DQB typing was carried out in two wells. Each well contained one primer pair and two probes labeled either with FAM™ (6-carboxyfluorescein) or VIC® and modified with a minor groove binder (MGB) and a non-fluorescent quencher (Applied Biosystems, Inc.). These primer sets and probes distinguish the major DQB polymorphisms at codons 25–28 and 47–48. In combination, the primers and probes identify the allele DQB1*0602 and distinguish several additional DQB1 alleles, as shown in Table 2. The primers and probes were designated as GDQ1 through GDQ8 (Genotyping oligonucleotides for HLA-DQ), as shown in Table 1. Primers and probe for HLA-DRA (GDR1-3) were used in a third reaction well to control for template quality and quantity, as previously described (23).
Table 1.
Oligonucleotide sequences for HLA typing of DQB1*0602.
| Oligo Name | Description | Sequence (5′ to 3′) | Final concentration (nM) |
|---|---|---|---|
| DQB1 typing: | |||
| GDQ1 | DQB forward 132 primer | GGGCATGTGCTACTTCACCAA | 225 |
| GDQ2 | DQB reverse 209 primer | GCGTACTCCTCTCGGTTATAGATGT | 225 |
| GDQ3 | DQB 167 probe 1 | FAM-TGCGTCTTGTGACCAG-MGB | 50 |
| GDQ4 | DQB 167 probe 2 | VIC-TGCGTCTTGTAACCAG-MGB | 50 |
| GDQ5 | DQB forward 155 primer | GGACGGAGCGCGTGCGTCTT | 225 |
| GDQ6 | DQB reverse 294 primer | TTCCTTCTGGCTGTTCCAGTACT | 225 |
| GDQ7 | DQB 245 probe 1 | FAM-ACCGCGCGGTACA-MGB | 50 |
| GDQ8 | DQB 246 probe 2 | VIC-CACCGCCCGATAC-MGB | 50 |
| DRA control: | |||
| GDR1 | DRA forward primer | AGGCCGAGTTCTATCTGAATCCT | 225 |
| GDR2 | DRA reverse primer | CGCCAGACCGTCTCCTTCT | 225 |
| GDR3 | DRA probe | VIC-CATAAACTCGCCTGATTG-MGB | 50 |
Table 2.
Interpretation of QPCR-SSPP results for common DQB1 alleles.
| DQB1* | Probes |
||||
|---|---|---|---|---|---|
| GDR3 | GDQ3 | GDQ4 | GDQ7 | GDQ8 | |
| 0601 | + | ||||
| 0602 | + | + | + | ||
| 0603 | + | + | + | ||
| 060401 | + | + | |||
| 060402 | + | + | + | ||
| 060403 | + | + | |||
| 0605 | + | + | |||
| 0606 | + | + | |||
| 0607 | + | + | + | ||
| 060801 | + | + | + | ||
| 060802 | + | + | |||
| 0609 | + | + | |||
| 0610 | + | + | + | ||
| 061101 | + | + | + | ||
| 061102 | + | + | + | ||
| 0301 | + | ||||
| 0302 | + | + | + | ||
| 0303 | + | + | + | ||
| 0304 | + | ||||
| 0305 | + | ||||
| 0306 | + | + | + | ||
| 0307 | + | + | |||
| 0308 | + | + | + | ||
| 0309 | + | ||||
| 0310 | + | ||||
| 02 | + | ||||
| 04 | + | ||||
| 05 | + | ||||
“+” indicates that the probe gives a positive signal for this allele
Real-time amplification
Reactions were performed with approximately 30–50 ng of genomic DNA in TaqMan® Genotyping Master Mix (Applied Biosystems, Inc.) in a final volume of 25 microliters per well. All primers and probes were used at the final concentrations listed in Table 1. Reactions were amplified on an ABI PRISM® 7000 Sequence Detection System (Applied Biosystems, Inc.) as follows: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec, 60°C for 1 min. Samples were plated in singlate reactions in three wells. Well 1 contained control primers and probes GDR 1–3, well 2 contained oligonucleotides GDQ 1–4, and well 3 contained GDQ 5–8.
Analysis and interpretation
Thresholds (to measure fluorescence during the exponential phase of the PCR) were set at 0.05, except for GDQ3, which was set at 0.1, based on calibration with samples of known genotype. The cycle during which each sample reached this threshold level of fluorescence (CT) was determined. Reactions were scored as positive if the CT for GDR3 (DRA) was no more than 36 and the CT for GDQ probes was within 3 cycles (GDQ3 and GDQ4 ) or 6 cycles (GDQ7 and GDQ8) of the CT of GDR3. All algorithms were combined into a Microsoft® Office Excel (Microsoft Corp.) spreadsheet for rapid analysis of subtyping results based on the amplification patterns detailed in Table 2. DQB1*0602 was identified and differentiated from all common DQB1*06 alleles, although the rare alleles, DQB1*0610, *061101, *0613, *0615, *0616, *0620, *0624, *0629 and *0633, were not resolved. As shown in Table 2, some additional common DQB1*03 alleles (including DQB1*0302 and *0303, but excluding *0301) and other DQB1*06-non-DQB1*0602 alleles (e.g. DQB1*0603) were also identified by this assay.
Dose determination
Copy numbers of DQB1*0602, common DQB1*03, and other DQB1*06 alleles were determined by comparing the CT of GDR3 (DRA) with the CT of GDQ3, GDQ4, GDQ7, and GDQ8. Samples were scored as having two copies of a GDQ3-positive allele (either DQB1*0602 and/or *03, as shown in Table 2) if the dCT (CTGDQ3 - CTGDR3) was less than 1. Likewise, probes GDQ4, 7, and 8 were scored with two copies of detectable alleles (refer to Table 2) if the dCT (CTGDQx - CTGDR3) was less than 1, 2, or 1.7, respectively.
Results
Validation with blinded samples
To assess the reliability and accuracy of this typing strategy, 76 samples that had been previously typed using standard techniques (PCR-SSO, PCR-SSP, or PCR-SBT) were analyzed using the QPCR-SSPP rapid assay. Thirty-two were analyzed without blinding, and forty-four were blinded. Twenty-three of the seventy-six samples were accurately identified as positive for DQB1*0602, including six that were correctly identified as homozygous for this allele. Thirteen DQB1*06 alleles (other than DQB1*0602) were identified and discriminated from DQB1*0602. This included correct typing of three DNA samples that were heterozygous for DQB1*0602-DQB1*0603. Twenty-one samples were correctly typed as positive for DQB1*03. There were three discordancies between previous reference typing and the results of the QPCR-SSPP assay. Two of the samples had been reference typed by PCR-SSO as positive for DQB1*0603 but were found by QPCR-SSPP to be positive, instead, for DQB1*0602; one other sample that had been previously typed as positive for DQB1*0603 was shown to be negative for any DQB1*06 by QPCR-SSPP. To resolve these discrepancies, these samples were reanalyzed by sequence-based typing (PCR-SBT). In all three cases, the QPCR-SSPP typing was concordant with PCR-SBT results. Neither false positives nor false negatives were observed by QPCR-SSPP.
Determination of allele copy number
QPCR-SSPP can also be used to assess the number of copies of a specific allele, as described in Gersuk and Nepom (23). This ability to determine copy number relies on one of the capabilities of real-time PCR, in that the CT (the cycle at which accumulated fluorescence reaches a set threshold level) is proportional to the amount of starting material, and further, that a two-fold difference in copy number can be reliably differentiated. We have chosen the essentially non-polymorphic gene, HLA-DRA, as a reference gene to measure the amount of genomic DNA present. Since DRA is present in two copies in every individual, it is possible to compare the CT of other products to that of DRA to assess whether an individual carries one or two alleles of a particular type, in this case DQB1*0602, as well as the other alleles detected by this assay.
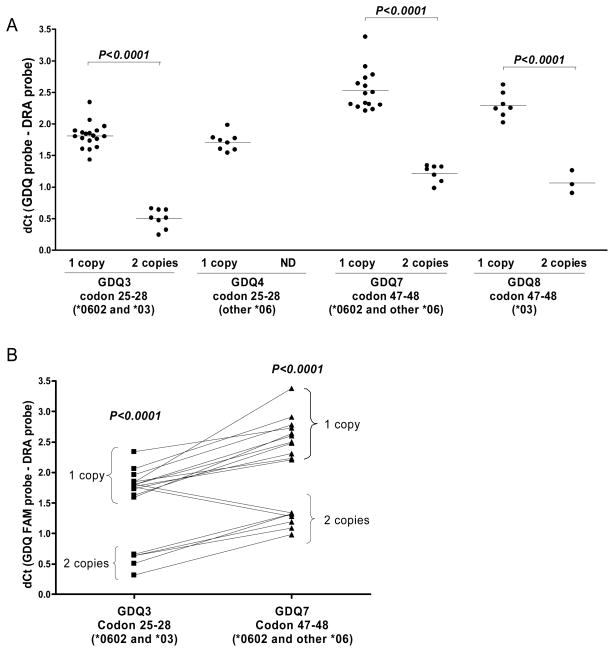
We tested all four GDQ probes for the ability to distinguish one copy from two copies. As shown in Figure 1A, the dCT (the CT from each GDQ probe compared to the CT of DRA) consistently distinguished the copy number, and in conjunction with Table 2, could be used to assess the dose of DQB1*0602, *03, and other *06 alleles in each sample. Homozygotes for DQB1*0602 were easily identified and distinguished from samples with only a single copy of DQB1*0602 by assessing copy number with probes GDQ3 and GDQ7, since only DQB1*0602 homozygotes display two dose equivalents with both of these probes (see Figure 1B). By evaluating copy number, along with the presence or absence of signal from each probe, several DQB1 genotypes could be distinguished (see Table 2). Further, since a positive determination of DQB1*0602 requires a positive result for two separate GDQ probes in two separate reaction wells (GDQ3 and GDQ7), along with the control GDR probe in a third well, the likelihood of false positives is minimized.
Figure 1.
Dose determination of DQB1 alleles. A. The dCT (CTGDQx - CTGDR3) for each GDQ probe was assessed in thirty-two previously typed DNA samples. Each dot represents one DNA sample identified by the indicated probe. The means of the dCT values for samples containing one- or two-copies of the identified alleles are indicated, and the differences were significant for GDQ3, 7, and 8 by a two-tailed, unpaired t test (p<0.0001). ND, there were no tested samples with two copies of a GDQ4-positive allele. An abbreviated key to the alleles identified by each probe is indicated in parentheses (see Table 2 for a complete key). B. DNA samples with a positive signal for probes GDQ3 and GDQ7 are displayed. Each line connects the dCT results for a single DNA sample for each of these two probes. Samples homozygous for DQB1*0602 are those that display two copies for both probes.
Reproducibility and assay variation
The DQB1*0602 QPCR-SSPP assay uses multiple built-in control elements, including simultaneous amplification of DRA as a control for quality and quantity and the use of pairs of single- and multiple-nucleotide polymorphism-detecting probes for subtype determination. These control elements provide internal checks that make running the reactions in singlate feasible. In developing the QPCR-SSPP assay, a subset of both blinded and unblinded samples were tested in replicate, both on the same assay plate and on separate plates. Identification of HLA subtype was found to be equally accurate with replicates, run separately or simultaneously, as with singlate reactions (data not shown). The use of singlate reactions resulted in increased throughput without a loss of accuracy. This high throughput allowed us to evaluate a large cohort of 734 individuals, including patients diagnosed with narcolepsy along with unaffected controls, for a study of the prevalence of narcolepsy in King County, WA (25). Using DNA isolated from buccal swabs, we observed 238 samples that were positive for DQB1*0602, including 143 samples positive for a single copy of DQB1*0602, thirty-five that were homozygous for DQB1*0602, plus, twenty-one that were positive for DQB1*0602 and another *06 allele, and thirty-nine that were positive for DQB1*0602 and *03.
Discussion
HLA-DQB1*0602 is associated with susceptibility to or protection from several diseases, so typing for this allele can be an important component of many clinical and research studies. Yet there is not a reliable, cost-effective method for directed identification. We wished to evaluate a large narcolepsy cohort of 734 subjects for the presence and copy number of the closely-associated risk allele, DQB1*0602. Using standard techniques would have been prohibitive in terms of both effort and cost. For this reason, we set out to develop a targeted DQB1*0602 assay, based on the previously described QPCR-SSPP TaqMan-based typing technique (23). The QPCR-SSPP technique can be applied to both targeted allele identification, as described here, as well as routine high and low resolution HLA typing. It can be also be used to rapidly determine allele copy number (zygosity). The approach is sensitive, specific and accurate.
Targeted typing for DQB1*0602 is complex because of the number and distribution of polymorphisms in the DQB1 locus. However, by using a combination of primers and probes that differentiate alleles based on these polymorphisms, we designed an assay that is performed in a total of only three reaction wells—two for the identification of DQB1*0602 and one for quantity and quality control. Thus, identification of DQB1*0602 can be performed on 32 samples at a time in a typical 96-well format plate. With standard thermocycler speeds, a run takes approximately 1½ hours, most of which is hands-free. We have conducted limited testing on a fast-block QPCR platform, which allows 40 minute run times, and have shown that this assay can be successfully adapted to such a rapid system (data not shown). Thus, using a 384-well and/or fast-block platform, throughput, time, and cost savings could all be further enhanced. Most of the effort required by the assay is automated and easily adaptable to robotic devices. The assay is performed in a sealed plate and no post-PCR manipulation is required. To simplify analyses, we have created a spreadsheet in Microsoft® Excel, containing algorithms that quickly analyze data and generate typing results automatically (available from the authors on request). Further, the assay allows for the determination of allele copy numbers, which can be an important component in evaluating disease risk, since both relative risk and severity can be affected by the presence and copy number of susceptibility alleles (4;26–29). Using standard HLA typing techniques, copy number is determined by inference, based on other alleles detected. In contrast, QPCR-SSPP measures actual copy number directly, providing a simple, inexpensive option for assessing zygosity.
A small number of recent reports describe the use of real-time PCR for HLA typing (e.g. (23;30–34)). We have shown here that HLA-DQB1*0602 typing and dose determination can be accomplished rapidly, accurately, and in a cost-efficient manner. This real-time PCR assay is especially useful for rapid targeted typing of large sample numbers in both research and clinical laboratories and further demonstrates the usefulness of real-time PCR approaches for HLA typing determinations.
Acknowledgments
This work was supported by grants from the Juvenile Diabetes Research Foundation (4-2007-1058), The National Institutes of Health (DK061036, AI050864), and the National Institute of Neurological Disorders and Stroke (NS038523). We thank Will Leighty and the BRI Clinical Core Registry for providing genomic DNA samples for validation analyses, Lakshmi Gaur and the Puget Sound Blood Center for reference HLA typing of some of the DNA samples, and William Longstreth for providing samples from his narcolepsy cohort.
Abbreviations
- HLA
Human leukocyte antigen
- MGB
Minor Groove Binder
- PCR
Polymerase chain reaction
- PCR-SBT
Polymerase chain reaction with sequence-based typing
- PCR-SSO
Polymerase chain reaction with sequence specific oligonucleotide probes
- PCR-SSP
Polymerase chain reaction with sequence specific primers
- QPCR-SSPP
Quantitative real-time PCR with sequence specific primers and probes
- MS
Multiple Sclerosis
- T1D
Type I Diabetes
References
- 1.Doherty D, Penzotti JE, Koelle DM, et al. Structural basis of specificity and degeneracy of T cell recognition: Pluriallelic restriction of T cell responses to a peptide antigen involves both specific and promiscuous interactions between the T cell receptor, peptide, and HLA-DR. Journal of Immunology. 1998;161:3527–35. [PubMed] [Google Scholar]
- 2.Gebe JA, Novak EJ, Kwok WW, Farr AG, Nepom GT, Buckner JH. T cell selection and differential activation on structurally related HLA-DR4 ligands. Journal of Immunology. 2001;167:3250–6. doi: 10.4049/jimmunol.167.6.3250. [DOI] [PubMed] [Google Scholar]
- 3.Nepom GT. The role of the DR4 shared epitope in selection and commitment of autoreactive T cells in rheumatoid arthritis. Rheumatic Disease Clinics of North America. 2001;27 (2):305–15. doi: 10.1016/s0889-857x(05)70203-9. [DOI] [PubMed] [Google Scholar]
- 4.Wordsworth P, Pile KD, Buckely JD, et al. HLA heterozygosity contributes to susceptibility to rheumatoid arthritis. Am J Hum Genet. 1992;51 (3):585–91. [PMC free article] [PubMed] [Google Scholar]
- 5.Chabas D, Taheri S, Renier C, Mignot E. The genetics of narcolepsy. Annu Rev Genomics Hum Genet. 2003;4:459–83. doi: 10.1146/annurev.genom.4.070802.110432. [DOI] [PubMed] [Google Scholar]
- 6.Mignot E, Hayduk R, Black J, Grumet FC, Guilleminault C. HLA DQB1*0602 is associated with cataplexy in 509 narcoleptic patients. Sleep. 1997;20 (11):1012–20. [PubMed] [Google Scholar]
- 7.Rogers AE, Meehan J, Guilleminault C, Grumet FC, Mignot E. HLA DR15 (DR2) and DQB1*0602 typing studies in 188 narcoleptic patients with cataplexy. Neurology. 1997;48 (6):1550–6. doi: 10.1212/wnl.48.6.1550. [DOI] [PubMed] [Google Scholar]
- 8.Hillert J, Olerup O. HLA and MS. Neurology. 1993;43 (11):2426–7. doi: 10.1212/wnl.43.11.2426-a. [DOI] [PubMed] [Google Scholar]
- 9.Hong SC, Hayduk R, Lim J, Mignot E. Clinical and polysomnographic features in DQB1*0602 positive and negative narcolepsy patients: results from the modafinil clinical trial. Sleep Med. 2000;1 (1):33–9. doi: 10.1016/s1389-9457(99)00007-6. [DOI] [PubMed] [Google Scholar]
- 10.Arnett F, Reveille JD. Genetics of systemic lupus erythematosus. In: Nepom GT, editor. Rheumatic Disease Clinics of North America. Philadelphia: W.B. Saunders Company; 1992. pp. 865–92. [PubMed] [Google Scholar]
- 11.Arnett FC, Reveille JD, Moutsopoulos HM, Georgescu L, Elkon KB. Ribosomal P autoantibodies in systemic lupus erythematosus. Frequencies in different ethnic groups and clinical and immunogenetic associations. Arthritis and Rheumatism. 1996;39 (11):1833–9. doi: 10.1002/art.1780391109. [DOI] [PubMed] [Google Scholar]
- 12.Cortes LM, Baltazar LM, Lopez-Cardona MG, et al. HLA class II haplotypes in Mexican systemic lupus erythematosus patients. Hum Immunol. 2004;65 (12):1469–76. doi: 10.1016/j.humimm.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Voorter CE, Drent M, van den Berg-Loonen EM. Severe pulmonary sarcoidosis is strongly associated with the haplotype HLA-DQB1*0602-DRB1*150101. Hum Immunol. 2005;66 (7):826–35. doi: 10.1016/j.humimm.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Spurkland A, Saarinen S, Boberg KM, et al. HLA class II haplotypes in primary sclerosing cholangitis patients from five European populations. Tissue Antigens. 1999;53 (5):459–69. doi: 10.1034/j.1399-0039.1999.530502.x. [DOI] [PubMed] [Google Scholar]
- 15.Apple RJ, Erlich HA, Klitz W, Manos MM, Becker TM, Wheeler CM. HLA DR-DQ associations with cervical carcinoma show papillomavirus-type specificity. Nature Genetics. 1994;6 (2):157–62. doi: 10.1038/ng0294-157. [DOI] [PubMed] [Google Scholar]
- 16.Apple RJ, Becker TM, Wheeler CM, Erlich HA. Comparison of human leukocyte antigen DR-DQ disease associations found with cervical dysplasia and invasive cervical carcinoma. J Natl Cancer Inst. 1995;87 (6):427. doi: 10.1093/jnci/87.6.427. [DOI] [PubMed] [Google Scholar]
- 17.Quintero E, Pizarro MA, Rodrigo L, et al. Association of Helicobacter pylori-related distal gastric cancer with the HLA class II gene DQB10602 and cagA strains in a southern European population. Helicobacter. 2005;10 (1):12–21. doi: 10.1111/j.1523-5378.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- 18.Sanjeevi CB, Hjelmstrom P, Hallmans G, et al. Different HLA-DR-DQ haplotypes are associated with cervical intraepithelial neoplasia among human papillomavirus type-16 seropositive and seronegative Swedish women. Int J Cancer. 1996;68 (4):409–14. doi: 10.1002/(SICI)1097-0215(19961115)68:4<409::AID-IJC1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 19.Pugliese A, Gianani R, Moromisato R, et al. HLA-DQB1*0602 is associated with dominant protection from diabetes even among islet cell antibody-positive first-degree relatives of patients with IDDM. Diabetes. 1995;44 (6):608–13. doi: 10.2337/diab.44.6.608. [DOI] [PubMed] [Google Scholar]
- 20.Saiki RK, Bugawan TL, Horn GT, Mullis KB, Erlich HA. Analysis of enzymatically amplified beta-globin and HLA-DQ alpha DNA with allele-specific oligonucleotide probes. Nature. 1986;324 (6093):163–6. doi: 10.1038/324163a0. [DOI] [PubMed] [Google Scholar]
- 21.Olerup O, Zetterquist H. HLA-DR typing by PCR amplification with sequence-specific primers (PCR-SSP) in 2 hours: an alternative to serological DR typing in clinical practice including donor-recipient matching in cadaveric transplantation. Tissue Antigens. 1992;39 (5):225–35. doi: 10.1111/j.1399-0039.1992.tb01940.x. [DOI] [PubMed] [Google Scholar]
- 22.Sayer D, Whidborne R, Brestovac B, Trimboli F, Witt C, Christiansen F. HLA-DRB1 DNA sequencing based typing: an approach suitable for high throughput typing including unrelated bone marrow registry donors. Tissue Antigens. 2001;57 (1):46–54. doi: 10.1034/j.1399-0039.2001.057001046.x. [DOI] [PubMed] [Google Scholar]
- 23.Gersuk VH, Nepom GT. A real-time PCR approach for rapid high resolution subtyping of HLA-DRB1*04. J Immunol Methods. 2006;317 (1–2):64. doi: 10.1016/j.jim.2006.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Robinson J, Waller MJ, Parham P, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31 (1):311–4. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Longstreth WT, Jr, Ton TG, Koepsell TD, Gersuk VH, Hendrickson AF, Velde S. Sleep Medicine. Washington, USA: 2008. Prevalence of Narcolepsy in King County. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fries JF, Wolfe F, Apple R, et al. HLA-DRB1 genotype associations in 793 white patients from a rheumatoid arthritis inception cohort: frequency, severity, and treatment bias. Arthritis and Rheumatism. 2002;46 (9):2320–9. doi: 10.1002/art.10485. [DOI] [PubMed] [Google Scholar]
- 27.Mignot E, Lin L, Rogers W, et al. Complex HLA-DR and -DQ interactions confer risk of narcolepsy-cataplexy in three ethnic groups. Am J Hum Genet. 2001;68 (3):686–99. doi: 10.1086/318799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nepom GT, Hansen JA, Nepom BS. The molecular basis for HLA class II associations with rheumatoid arthritis. J Clin Immunol. 1987;7:1–7. doi: 10.1007/BF00915418. [DOI] [PubMed] [Google Scholar]
- 29.Nepom GT, Nepom BS. Prediction of susceptibility to rheumatoid arthritis by human leukocyte antigen genotyping. In: Nepom GT, editor. Rheumatic Disease Clinics of North America. Philadelphia: W.B. Saunders Company; 1992. pp. 785–92. [PubMed] [Google Scholar]
- 30.Casamitjana N, Faner R, Santamaria A, et al. Development of a new HLA-DRB real-time PCR typing method. Hum Immunol. 2005;66 (1):85–91. doi: 10.1016/j.humimm.2004.08.178. [DOI] [PubMed] [Google Scholar]
- 31.Reinton N, Helgheim A, Shegarfi H, Moghaddam A. A one-step real-time PCR assay for detection of DQA1*05, DQB1*02 and DQB1*0302 to aid diagnosis of celiac disease. J Immunol Methods. 2006;316 (1–2):125–32. doi: 10.1016/j.jim.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 32.Slateva K, Elsner HA, Albis-Camps M, Blasczyk R. HLA-DRB fluorotyping by dark quenching and automated analysis. Tissue Antigens. 2001;58 (4):250–4. doi: 10.1034/j.1399-0039.2001.580405.x. [DOI] [PubMed] [Google Scholar]
- 33.Sylvain K, Aurelie H, Marc M, Christophe R. Rapid screening for HLA-B27 by a TaqMan-PCR assay using sequence-specific primers and a minor groove binder probe, a novel type of TaqMan trade mark probe. J Immunol Methods. 2004;287 (1–2):179–86. doi: 10.1016/j.jim.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 34.Tremmel M, Opelz G, Mytilineos J. High-resolution typing for HLA-DRB1*15 and -DRB1*16 by fluorescence-marked sequence-specific priming (TaqMan assay) Tissue Antigens. 1999;54 (5):508–16. doi: 10.1034/j.1399-0039.1999.540508.x. [DOI] [PubMed] [Google Scholar]