Fig. 2.
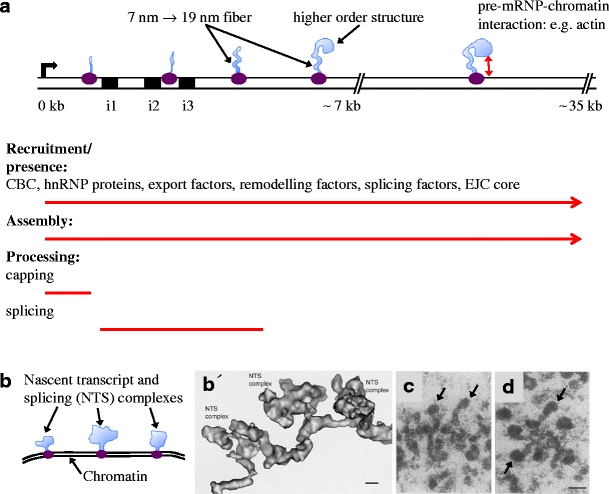
Cotranscriptional synthesis, assembly and processing of BR1/BR2 and BR3 pre-mRNPs. a The proximal (0–7 kb) and middle (7–35 kb) regions of the BR1/BR2 gene is shown schematically. The bent arrow indicates the transcription start site. The RNA polymerase II elongation complex in purple and the pre-mRNPs in light blue. The proximal region contains three introns (i1–i3, black boxes) and exons 1–3, followed by the long (approximately 35 kb) exon 4. The continuous recruitment of components to the pre-mRNPs (listed components have been demonstrated) and the continuous assembly of the pre-mRNPs are indicated by red arrows. The processing events of the pre-mRNPs are indicated (red lines). Assembly of the exon sequences results in a 7-nm pre-mRNP fibre that is further folded into a 19-nm fibre, followed by organisation into higher order structures. Interaction between the pre-mRNP and chromatin may involve pre-mRNP-bound actin. b Schematic representation of part of the BR3 gene. The BR3 gene contains multiple short introns and exons. Three nascent transcript and splicing (NTS) complexes are shown. Each complex consists of a pre-mRNP (light blue), including splicing factors and an RNA polymerase II elongation complex (purple). b′ EM 3D reconstruction of a corresponding BR3 gene segment with three NTS complexes (adopted from Wetterberg et al. 2001). The repeated assembly and structural dynamics of the spliceosome along the multi-intron gene dominate the structure of the pre-mRNPs. The scale bar represents 10 nm. c, d EM images of nascent BR1/BR2 pre-mRNPs in the middle region of the gene (adopted from Skoglund et al. 1983). The 19-nm fibre and the higher order granular structure are seen in several pre-mRNPs. The granular structure (arrows) gets bigger and denser as more RNA protein is added (compare c and d). The scale bar represents 50 nm
