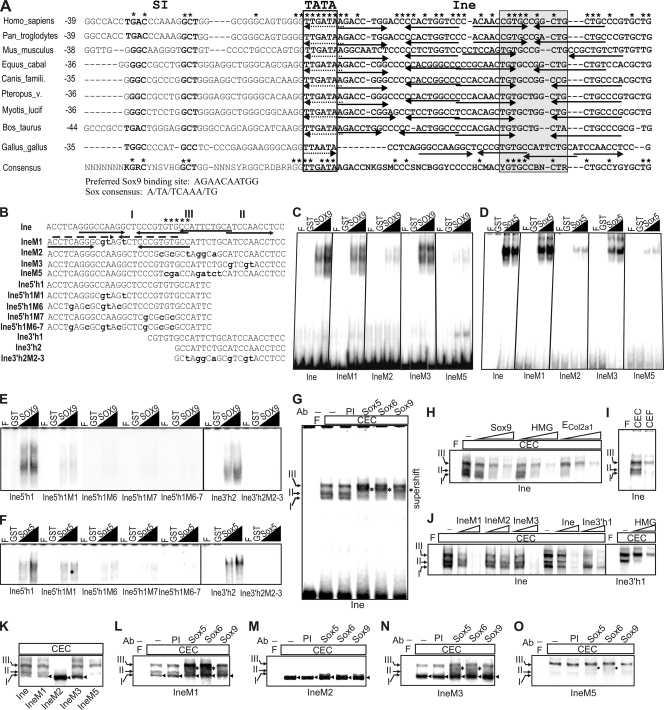
FIG. 3.
Binding of Sox proteins to Ine in vitro. (A) Ine sequences of selected amniotes and the 75% majority rule consensus for amniotes (see the whole alignment in Fig. S2A at http://www.brc.hu/pub/Supplemental_Material_Nagy_et_al_MCB2010.pdf). TATA and the most conserved motif are boxed; NFI motifs are in boldface. Positions are given from TATA. Nucleotides fully conserved in mammals or in amniotes are marked by asterisks at the top and above the consensus sequence, respectively. Arrows and dotted arrows depict motifs similar to the preferred Sox9-binding site (29) and Sox consensus (21), respectively. Equus_cabal, Equus caballus; Canis_famili., Canis familiaris; Pteropus_v., Pteropus vampyrus; Myotis_lucif, Myotis lucifugus. (B) Sequences of Ine and its shorter or mutant derivatives. The conserved GTGCC motif, mutant nucleotides, the 5′ (I) and 3′ (II) paired Sox sites and unrelated factor-binding site (III) are denoted. (C to F) EMSA of nucleoprotein complexes formed with purified GST-fused Sox proteins on Ine and its derivatives. (G to O) Binding of CEC nuclear proteins to Ine and Ine mutants. (G) Supershift analysis with Sox antibodies (Ab). (H) Competition EMSA with 50-, 100-, and 500-fold molar excesses of the indicated cold competitors. (I) Comparison of CEC and CEF nucleoprotein complexes. (J) Competition EMSA on Ine and Ine3′h1 with the cold probes indicated. (K to O) EMSA (K) and supershifts (L to O) of the wild-type and mutant Ine. The supershifts (asterisks) and new complexes of Ine mutants (arrowheads) are marked. F, free probe; PI, preimmune serum.