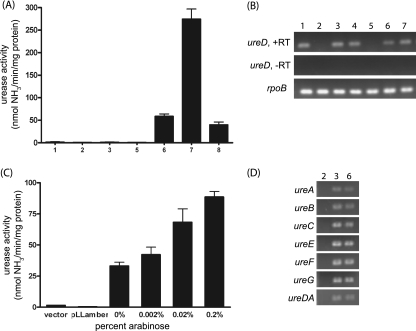
FIG. 1.
Urease activity and RT-PCR results for the Sakai ureD mutants. (A, B, and D) Lanes: 1, Sakai; 2, Sakai Δure; 3, Sakai ureD(Q); 4, Sakai Δure attTn7::ure (chromosomal complement of Sakai Δure; unpublished work); 5, E. coli MG1655; 6, EHEC IN1; 7, EHEC Mo28; 8, Sakai ureD(P,Q). (A) Cultures were grown overnight in DMEM supplemented with NiCl2, and the urease activities of cell lysates were measured. EHEC strains IN1 and Mo28 were used as positive controls, and MG1655 was used as a negative control. (B) RNA from overnight cultures was extracted and used for ureD RT-PCR experiments. (C) Sakai ureD(Q) harboring empty plasmid, pLLamber, or pLLQ was grown overnight, diluted 1:100 in M9 minimal medium supplemented with NiCl2 and ampicillin, grown to an OD600 of 0.45, induced with arabinose, and then grown for an additional 4 h. The urease activities of the Sakai ureD(Q) cell lysates were measured. The urease activity in lysates of Sakai ureD(Q) harboring pLLamber (the Sakai wild-type ureD) remained negligible at all concentrations of arabinose; thus, only that for 0.2% is included. (D) RNA from overnight cultures was extracted and used for detecting the transcript by RT-PCR.