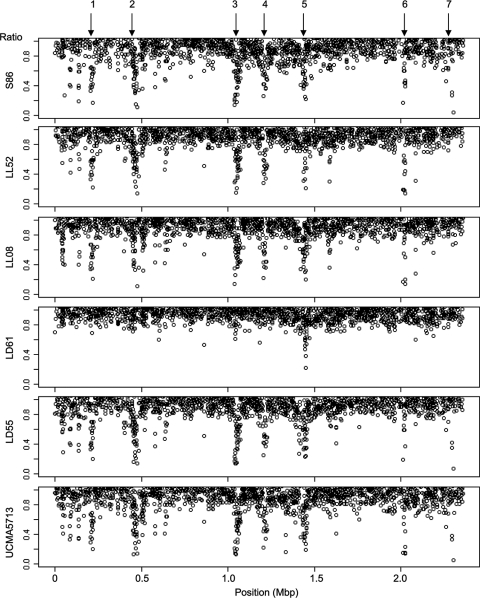
FIG. 2.
CGH results normalized using likely conserved genes and expressed as ratios of the signal intensities of the studied strains against that of the reference strain, IL1403 (statistical method c; see Table S1 in the supplemental material). Ratios lower than 1 for the six strains were plotted as a function of gene position on the IL1403 chromosome. Locus 1, position ∼0.20 Mbp, ycbABCDKFGHIJ and ycaFG genes; locus 2, position ∼0.44 Mbp, pi1 genes; locus 3, position ∼1.03 Mbp, pi2 genes; locus 4, position ∼1.20 Mbp, citRCDEF, mae, ymbCDHIJK, and ymcABCDEF genes; locus 5, position ∼1.41 Mbp, pi3 genes; locus 6, position ∼2.01 Mbp, ps3 genes; locus 7, position ∼2.30 Mbp, pspAB and ywjABCDEFGH genes. Genes were declared divergent only when their ratios were lower than 0.76.