Abstract
The influenza A virus polymerase complex, consisting of the subunits PB1, PB2, and PA, represents a promising target for the development of new antiviral drugs. We have previously demonstrated the feasibility of targeting the protein-protein interaction domain between PA and PB1 using peptides derived from the extreme N terminus of PB1 (amino acids [aa] 1 to 15), comprising the PA-binding domain of PB1. To increase the binding affinity of these peptides, we performed a systematic structure-affinity relationship analysis. Alanine and aspartic acid scans revealed that almost all amino acids in the core binding region (aa 5 to 11) are indispensable for PA binding. Using a library of immobilized peptides representing all possible single amino acid substitutions, we were able to identify amino acid positions outside the core PA-binding region (aa 1, 3, 12, 14, and 15) that are variable and can be replaced by affinity-enhancing residues. Surface plasmon resonance binding studies revealed that combination of several affinity-enhancing mutations led to an additive effect. Thus, the feasibility to enhance the PA-binding affinity presents an intriguing possibility to increase antiviral activity of the PB1-derived peptide and one step forward in the development of an antiviral drug against influenza A viruses.
Influenza A viruses cause respiratory febrile illness in humans claiming 250,000 to 500,000 lives annually (18). Incomplete protection by vaccines and the emergence of resistance to current antiviral drugs call for new strategies to inhibit influenza viruses. The polymerase complex, which consists of the three subunits PA, PB1, and PB2, has become an attractive target for the development of novel antivirals (2, 6, 23), including antivirals that block the assembly of the trimeric polymerase complex and thus viral transcription and replication (5, 8, 13, 22). Direct biochemical interactions have been shown for PB1 and PB2, as well as for PA and PB1 (1, 14-16, 19), whereas a weak transient interaction has been proposed for PA and PB2 (9). Such protein interaction interfaces are potential targets for the development of pharmaceutical inhibitors, including peptides that efficiently disrupt such protein-protein interactions. However, protein-protein interfaces frequently contain large surface areas, which make the successful development of suitable drugs a challenging task. In the case of the influenza virus polymerase complex, the N-terminal domain of PB1 (PB1N) interacts with the C-terminal domain of PA (PAC). Crystal structures have shown that the core of the PB1 interaction interface consists of only five residues (Pro5, Leu7, Leu8, Phe9, and Leu10) in a 310-helix (8, 13). Based on this core-binding domain, it has been speculated that the development of an antiviral peptide or peptidomimetic is feasible (8, 13, 17), especially in the light of a recently identified affinity-enhancing amino acid substitution in this binding domain (22) that might significantly improve the antiviral activity.
We recently provided evidence that peptides of 25 amino acids (aa), PB11-25, of PB1 efficiently bound to PA and showed antiviral activity against influenza A viruses by disrupting the PB1-PA interaction (5). We then further demonstrated that binding to PA was preserved with peptides of 15 aa in length (PB11-15) and that an enhanced binding affinity of a PB1-derived peptide correlates with increased antiviral activity (22). However, direct proof that the core PA-binding domain of PB1 (PB15-11) can bind to PA efficiently is missing. Consequently, we wanted to clarify (i) whether peptides only comprising the core PA-binding domain can be used as lead peptides for drug development and (ii) whether we could further improve the binding affinity of the PB1-derived peptide to the PA protein.
Based on a comprehensive structure-affinity-relationship analysis, we now show that the described core-binding region of PB1 (PB15-11) is not sufficient for PA binding and should be extended to aa 2 to 12. Importantly, our data suggest that increased PA binding of PB11-15 can be achieved by affinity enhancing mutations. Four of five identified affinity-enhancing mutations are located outside the core-binding region, confirming the important role of amino acids outside of this region. Furthermore, combinations of the affinity-enhancing amino acid substitutions resulted in a high-affinity peptide.
MATERIALS AND METHODS
Peptide synthesis.
The solid-phase synthesis of the peptides was carried out on a Pioneer automatic peptide synthesizer (Applied Biosystems, Foster City, CA) using Fmoc (9-fluorenylmethoxy carbonyl) chemistry with O-(benzotriazol-1-yl)-N,N,N′,N′,-tetramethyluronium tetrafluoroborate (TBTU)/diisopropylethyl amine activation. Side chain protections were as follows: for Asp, Glu, Ser, Thr, and Tyr: t-Bu; Asn, Gln, and His, Trt was used; for Arg, Pbf was used; and for Lys and Trp, Boc was used. The coupling time was 1 h. Double couplings were carried out if a difficult coupling was expected according to the program Peptide Companion (CoshiSoft/PeptiSearch, Tucson, AZ). All peptides were generated as carboxyl amides by synthesis on Rapp S RAM resin (Rapp Polymere, Tübingen, Germany). Biotin was incorporated at the C termini of the indicated peptides with Fmoc-Lys(Biotin)-OH (NovaBiochem/Merck, Nottingham, United Kingdom) and TBTU/diisopropylethyl amine activation for 18 h, followed by coupling of Fmoc-b-Ala-OH for 1 h. Peptides were cleaved from the resin and deprotected by a 3-h treatment with trifluoroacetic acid (TFA) containing 3% triisobutylsilane and 2% water (10 ml/g resin). After precipitation with t-butylmethyl ether, the resulting crude peptides were purified by preparative high-pressure liquid chromatography (HPLC; RP-18) with water-acetonitrile gradients containing 0.1% TFA and characterized by analytical HPLC and matrix-assisted laser desorption ionization-time of flight mass spectroscopy. Some peptides were synthesized by Peptides&elephants GmbH (Potsdam, Germany) and subsequently purified and characterized as described above.
ELISA.
Streptavidin-coated microwell plates were incubated with saturating concentrations of biotinylated peptides, washed, and subsequently incubated at room temperature with hemagglutinin (HA)-tagged PA. To obtain PA-HA, 293T cells were seeded into 94-mm dishes, transfected with the respective plasmid, and treated with lysis buffer at 24 h posttransfection as previously described (21). After the microwell plates were washed, the wells were incubated with an HA-specific primary antibody (MMS-101R; Covance), followed by three washes and an incubation with a peroxidase-coupled secondary antibody (Jackson Immunoresearch, Newmarket, United Kingdom). After the final wash step, ABTS [2,2′azinobis(3-ethylbenzthiazolinesulfonic acid)] substrate (Sigma Ready-to-Use solution) was added, and the optical density was determined at 405 nm. A competition enzyme-linked immunosorbent assay (ELISA) was carried out as described above except that the competitor peptides were added to wells of the plate with bound peptides simultaneously with the addition of the cell extract containing HA-tagged PA subunits.
Synthesis of peptide arrays and binding of PA.
Membranes with peptide arrays were synthesized by the previously described SPOT technique (4). Each array contained 300 15-mer peptides arranged in rows of 25 spots. Sequences are systematic single substitutions of the wild-type (wt) PB11-15 and the mutant PB11-15T6Y. Each array displays the parent peptide sequence 15 times for an internal quality control.
The peptides were synthesized by using Fmoc chemistry with N,N′ disopropylcarbodiimide (DIC) and HOBt-diisopropylethylamine activation. The same amino acid derivates as described for the peptide synthesis (see above) were used. Acid stable amino-PEGylated cellulose membranes (AIMS-Scientific Products GmbH, Braunschweig, Germany) were used. Double couplings were carried out, followed by a 2-h reaction time to elongate the peptide chains. Every elongation cycle was continued by an acetylation step with 2% acetic acid anhydride in dimethylformamide (DMF) to cap all unreacted free amino functions. Three washes with DMF, Fmoc deprotection with 20% piperidine in DMF, three DMF washes, staining of free amino groups with 1% bromophenol blue in DMF, three washes with ethanol, and drying completed every elongation cycle. The working up order after the last elongation cycle was changed to three washes with DMF, Fmoc deprotection with 20% piperidine in DMF, three more washes with DMF, bromophenol blue staining, terminal acetylation with acetylation mix, three washes with ethanol, and drying. For the final side chain deprotection, the membranes were treated overnight with a solution of 80% trifluoroacetic acid, 12% dichloromethane (DCM), 5% water, and 3% triisobutylsilane. After four washes with DCM, three washes with DMF, two washes with ethanol, three washes with 1 M acetic acid in water, and four washes with ethanol, the membranes were dried.
Before binding of PA to the peptides, the membrane was washed with ethanol to rehydrate some hydrophobic peptide spots. After three washes with Tris-buffered saline (TBS), the membranes were blocked overnight with a casein-based blocking buffer (Sigma-Genosys, Inc.). After one wash with T-TBS (TBS plus 0.05% Tween 20), the first incubation with 1:10-diluted PA-HA (A/WSN/33) in blocking buffer for 1 h was carried out. After three washes with T-TBS, the membranes were incubated with a 1:1,000-diluted monoclonal anti-HA antibody in blocking buffer for 1 h. After three washes with T-TBS, an alkaline phosphatase (AP)-conjugated goat anti-mouse IgG antibody, diluted 1:500 in blocking buffer, was used for 1 h as the detection antibody. After 15 min of development with an MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide]-BCIP (5-bromo-4-chloro-3-indolylphosphate) mixture as a coloring solution, the membrane was washed and scanned for analysis.
Purification of recombinant PA proteins from yeast.
To prevent proteolysis, all manipulations were carried out at 4°C. Yeast cells were harvested by centrifugation at 800 × g for 5 min and resuspended in disruption buffer (20 mM Na2HPO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole, 10% isopropanol, 2 mM phenylmethylsulfonyl fluoride). Protease inhibitor cocktail tablets (Roche) were added to the disruption buffer (1 tablet/20 g of wet yeast biomass). Cells were cyclically disrupted by 12 mixings with glass beads. The PA-His proteins containing the soluble protein fraction (SPF) were then separated by centrifugation at 3,500 × g for 10 min and filtered through a 0.45-μm-pore-size membrane (Millipore). For the purification of PA-His, separated SPF was mixed with Ni-NTA agarose (Qiagen), which was previously equilibrated in binding buffer (20 mM Na2HPO4 [pH 8.0], 300 mM NaCl, 20 mM imidazole), and incubated for 40 min in batch mode. Thereafter, the resin was loaded into the column and washed with 5 column volumes (CV) of binding buffer, 5 CV of wash solution 1 (20 mM Na2HPO4 [pH 7.4], 500 mM NaCl, 20 mM imidazole, 2% Tween 20, 1% glycerol), and finally 5 CV of wash solution 2 (20 mM Na2HPO4 [pH 7.4], 300 mM NaCl, 20 mM imidazole). Recombinant PA-His was eluted with elution buffer (20 mM Na2HPO4 [pH 7.4], 300 mM NaCl, and 250 mM imidazole) in a one-step gradient. The highest PA-His concentrations containing fractions were pooled, and imidazole was removed during a desalting step using a Millipore ultrafiltration device.
For the purification of PA-HA, collected yeast cells were disrupted as described before for PA-His with minor differences: the concentration of NaCl in the disruption buffer was reduced to 20 mM, and Tris-HCl was used as a buffer base instead of Na2HPO4. A separated SPF was run through a Q-Sepharose column (Tosoh), which was previously equilibrated in binding buffer (20 mM Tris-HCl [pH 8.0], 20 mM NaCl, 10% of glycerol), and PA-HA proteins were eluted by using an NaCl gradient (from 0.02 to 1 M) in 20 mM Tris-HCl (pH 8.0) and 10% glycerol. Next, collected elution fractions were analyzed by SDS-PAGE and tested for PA activity in an ELISA-based binding assay. The highest concentrations of active PA-HA containing fractions were pooled and analyzed for PA-HA purity by SDS-PAGE.
Surface plasmon resonance analysis.
All interaction studies were performed in real time with a BIAcore 3000 system (GE Healthcare). The experiments were done at 25°C at a flow rate of 30 μl min−1 in Tris-based buffer (25 mM Tris [pH 7.4], 150 mM NaCl, 0.01% surfactant). Peptides containing the N-terminal 15 aa of PB1 and a C-terminally fused biotin tag were immobilized on streptavidin sensor chip. A scrambled control peptide was used (which contained the same amino acids as PB11-25 but in a random order) (22). Association (1 min) of the purified, recombinant PA proteins was followed by a 2-min dissociation phase during which the used buffer was perfused. For analysis of the affinity constants, various concentrations of the analyte were injected. To regenerate the chip, the bound protein was removed by a 10-s pulse of 50 mM NaOH. The obtained data were analyzed with BIAevaluation 3.1 evaluation software using a 1:1 Langmuir binding model for calculation of the association and dissociation rate constants (Kon and Koff) and the dissociation equilibrium constants (KD = Koff/Kon).
RESULTS
Amino acids outside the core-binding region of PB1 are essential for PA binding.
To determine the minimal requirements of the core PA-binding domain of PB1 (PB15-11), which was previously proposed by structural analysis (8, 13), we measured the 50% inhibitory concentration (IC50) of this peptide in a competition ELISA (21). No detectable binding was observed with PB15-11 (Table 1). Furthermore, the binding affinity could not be enhanced by substitution of aa 6 with either Y (PB15-11T6Y) or W (PB15-11T6W), two previously described affinity-enhancing amino acid substitutions (21). In contrast, peptide PB11-11, harboring the four N-terminal amino acids, regained binding affinity, although the IC50 was 39-fold higher than with PB11-15.
TABLE 1.
Efficiency of PB1 peptides to compete with PA bindinga
| Peptide | Amino acid sequence | Mean IC50 (nM) ± SD |
|---|---|---|
| PB11-15 | MDVNPTLLFLKVPAQ | 41.3 ± 11.1 |
| PB11-11 | MDVNPTLLFLK---- | 1,611.3 ± 281.0 |
| PB15-11 | ----PTLLFLK---- | >10,000* |
| PB15-11T6Y | ----PYLLFLK---- | >10,000* |
| PB15-11T6W | ----PWLLFLK---- | >10,000* |
Determination of the 50% inhibitory concentrations (IC50s) of PB11-15-derived peptides by competitive ELISA using cell extract containing HA-tagged PA of A/WSN/33 and increasing concentrations of competitive peptides. Asterisks indicate the highest concentrations of competitive peptides (10,000 nM) used without detectable inhibitory effect. Affinity-enhancing amino acids are highlighted in boldface.
Identification of affinity-enhancing amino acid substitutions in PB11-15.
To enhance the affinity of PB11-15 for PA, we performed a structure-affinity relationship analysis as previously described for the antiviral peptide VIRIP (12) and an importin α peptide inhibitor (10). First, critical residues were determined by replacement of every single amino acid position in PB11-15 by either alanine or aspartic acid residues. Analysis of these PB11-15 derivatives in the competitive ELISA-based binding assay revealed the following results (Table 2). (i) The 310-helix of the core-PA-binding domain of PB1 that is constituted by the hydrophobic stretch P5TLLFLK11 does not tolerate any amino acid replacement by alanine (A) or aspartic acid (D), without losing binding affinity, except for the T6A substitution. Compared to the IC50 of PB11-15, this substitution reduces the affinity 45-fold. (ii) Residues outside the 310-helix can be substituted by A or D without complete loss of competitive activity. Nevertheless, these substitutions at positions 1 to 3 and positions 11 to 13 are all associated with an increase in IC50, indicating that substitutions by A or D are unfavorable for binding. (iii) Substitutions by D consistently display a more pronounced increase in IC50 than substitutions by A (compare, for example, PB11-15V12A to PB11-15V12D). This difference might be caused by the hydrophobic nature of the binding funnel provided by four α-helices in the C terminus of the PA protein (13), which might be disturbed by the negative charge of the D residues. Furthermore, A residues are more likely to preserve the helix structure of the peptide than D residues and thereby support binding affinity.
TABLE 2.
Efficiency of PB1 peptides carrying single alanine or aspartic acid substitutions to compete with PA bindinga
| Peptide | Amino acid sequence | Mean IC50 (nM) ± SD |
|---|---|---|
| PB11-15 | MDVNPTLLFLKVPAQ | 41.3 ± 11.1 |
| PB11-15M1A | ADVNPTLLFLKVPAQ | 596.7 ± 107.6 |
| PB11-15D2A | MAVNPTLLFLKVPAQ | 225.0 ± 63.4 |
| PB11-15V3A | MDANPTLLFLKVPAQ | 93.4 ± 31.8 |
| PB11-15N4A | MDVAPTLLFLKVPAQ | >3,000* |
| PB11-15P5A | MDVNATLLFLKVPAQ | >3,000* |
| PB11-15T6A | MDVNPALLFLKVPAQ | 1,835.7 ± 768.5 |
| PB11-15L7A | MDVNPTALFLKVPAQ | >3,000* |
| PB11-15L8A | MDVNPTLAFLKVPAQ | >3,000* |
| PB11-15F9A | MDVNPTLLALKVPAQ | >3,000* |
| PB11-15L10A | MDVNPTLLFAKVPAQ | >3,000* |
| PB11-15K11A | MDVNPTLLFLAVPAQ | 713.1 ± 140.9 |
| PB11-15V12A | MDVNPTLLFLKAPAQ | 427.9 ± 59.3 |
| PB11-15P13A | MDVNPTLLFLKVAAQ | 156.5 ± 51.1 |
| PB11-15M1D | DDVNPTLLFLKVPAQ | 703.5 ± 279.6 |
| PB11-15V3D | MDDNPTLLFLKVPAQ | >3,000* |
| PB11-15N4D | MDVDPTLLFLKVPAQ | >3,000* |
| PB11-15P5D | MDVNDTLLFLKVPAQ | >3,000* |
| PB11-15T6D | MDVNPDLLFLKVPAQ | >3,000* |
| PB11-15L7D | MDVNPTDLFLKVPAQ | >3,000* |
| PB11-15L8D | MDVNPTLDFLKVPAQ | >3,000* |
| PB11-15F9D | MDVNPTLLDLKVPAQ | >3,000* |
| PB11-15L10D | MDVNPTLLFDKVPAQ | >3,000* |
| PB11-15K11D | MDVNPTLLFLDVPAQ | >3,000* |
| PB11-15V12D | MDVNPTLLFLKDPAQ | 1,392.5 ± 330.7 |
| PB11-15P13D | MDVNPTLLFLKVDAQ | 414.6 ± 145.0 |
Determination of the IC50s of PB11-15 peptides harboring the indicated amino acid substitutions by competitive ELISA using increasing concentrations of competitive peptides and cell extract containing HA-tagged PA of A/WSN/33. Asterisks indicate the highest concentrations of competitive peptides (3,000 nM) used without detectable inhibitory effect. Amino acid replacements compared to the wt are indicated in boldface.
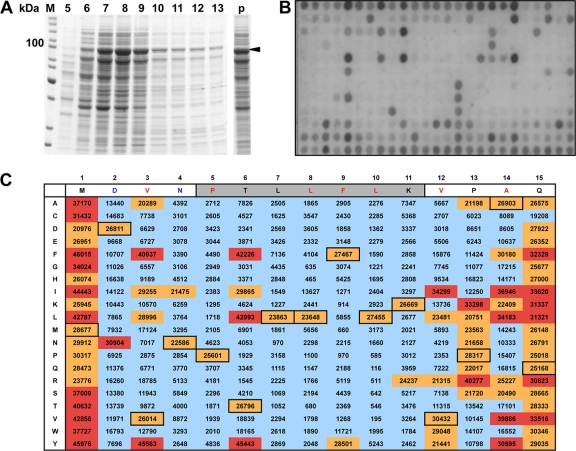
For a more comprehensive overview of the effect of mutations, we next generated and screened a peptide library of 300 PB11-15 variants, in which every single residue was individually substituted by all 20 natural amino acids. For this purpose, mutant peptides were spot synthesized on cellulose membranes (4) and tested for PA binding. Signals of PA-HA binding to the 15 copies of the parent peptide showed a relative mean value of 26,427 ± 2,050 (Fig. 1), proving the peptide array to be a reliable binding assay. Exchangeable amino acid positions were confined to aa 1 to 3, 6, and 13 to 15, confirming the results obtained with the A- and D-substituted peptides. For the nonexchangeable amino acid positions, we observed up to a 50-fold reduction in signal intensity compared to the mean value of the wt sequence. Interestingly, several affinity-enhancing mutations (red boxes) could be identified, which increased binding to values up to 45,976. In most cases, conservative substitutions resulted in enhanced binding: methionine (M) at position 1 could be replaced by a hydrophobic amino acid such as valine (V) or isoleucine (I) or an amino acid containing an aromatic ring structure such as tyrosine (Y) or phenylalanine (F) with a corresponding increase in binding affinity (Fig. 1). Aspartate at position 2 could only be substituted with asparagine (N), which most likely preserves the hydrogen bond to D412 of PA (13). At position 3, the substitutions of V to T or phenylalanine (F) resulted in a significant increase in PA binding. For position 6, we confirmed the affinity enhancing effect of replacing threonine (T) by either Y or F (22). Alanine (A) at position 14 mediates a hydrophobic contact to the PA protein and can be exchanged by similar amino acids such as isoleucine (I), leucine (L), V, or Y. Although proline (P) 13 and glutamine (Q) 15 do not show contact to PA in the crystal structure (13), substitutions with K and R (P13K and P13R) or F and V (Q15F and Q15V), respectively, resulted in enhanced binding to PA.
FIG. 1.
Binding characteristics of PA-HA to PB11-15 derivatives in an analogue peptide array. (A) Partial purification of PA-HA from yeast. C-terminally HA-tagged PA from A/WSN/33 was expressed in yeast, purified from the soluble fraction by anion-exchange chromatography via a Q-Sepharose column and analyzed for its purity by SDS-PAGE and subsequent Coomassie blue staining. Elution fractions 5 to 13 are shown. The arrow indicates the position of PA-HA. (B) Binding of PA-HA to immobilized PB11-15. An array with PB11-15 peptides harboring single amino acid substitutions as indicated in C was incubated with partially purified PA-HA (pooled fraction 6 to 9) from panel A. Bound PA-HA was detected by an HA-specific antibody and an AP-coupled secondary antibody. (C) Quantification of the signals on the peptide array in panel B. The PB11-15 wt sequence is shown horizontally; individual amino acid substitutions are shown vertically. Amino acids that mediate hydrogen bonds in the wt sequence are in blue letters; amino acids that mediate hydrophobic contacts are in red letters. The gray shading indicates the 310-helix. Numbers in black boxes represent wt peptide sequences, for which the mean is 26,427 with a standard deviation of 2,050. Quantifications of the signals of the array are categorized as <20,000 (blue shading), 20,000 to 30,500 (orange shading, including wt sequences), and >30,500 (red shading representing an increase in signal compared to wt sequences).
To confirm that the amino acid substitutions described above indeed increase the affinity to PA, we retested 11 selected peptides in the competitive ELISA and determined their IC50s (Table 3). Whereas only two peptides (PB11-15D2N and PB11-15Q15V) showed increased IC50s and thus no affinity-enhancing properties, the IC50s of all other peptides were 1.6- to 7-fold lower than that of PB11-15.
TABLE 3.
Efficiency of PB1 peptides carrying affinity-enhancing amino acid substitutions to compete with PA bindinga
| Peptide | Amino acid sequence | Mean IC50 (nM) ± SD |
|---|---|---|
| PB11-15M1Y | YDVNPTLLFLKVPAQ | 12.7 ± 3.7 |
| PB11-15D2N | MNVNPTLLFLKVPAQ | 45.2 ± 8.3 |
| PB11-15V3Y | MDYNPTLLFLKVPAQ | 6.3 ± 2.1 |
| PB11-15T6Y | MDVNPYLLFLKVPAQ | 21.64 ± 1.5 |
| PB11-15P13R | MDVNPTLLFLKVRAQ | 11.4 ± 1.7 |
| PB11-15P13K | MDVNPTLLFLKVKAQ | 16.6 ± 5.0 |
| PB11-15A14I | MDVNPTLLFLKVPIQ | 21.7 ± 9.3 |
| PB11-15A14L | MDVNPTLLFLKVPLQ | 36.3 ± 14.8 |
| PB11-15A14V | MDVNPTLLFLKVPVQ | 22.2 ± 5.4 |
| PB11-15Q15V | MDVNPTLLFLKVPAV | 52.2 ± 16.2 |
| PB11-15Q15F | MDVNPTLLFLKVPAF | 13.9 ± 2.8 |
Determination of the IC50s of PB11-15 peptides harboring the affinity-enhancing amino acid substitutions by competitive ELISA using cell extract containing HA-tagged PA of A/WSN/33 and increasing concentrations of competitive peptides. Amino acid replacements compared to the wt are indicated in boldface.
Combination of the single affinity enhancing mutations has an additive effect on affinity.
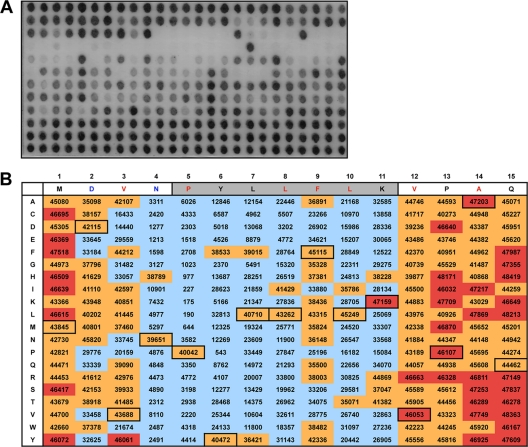
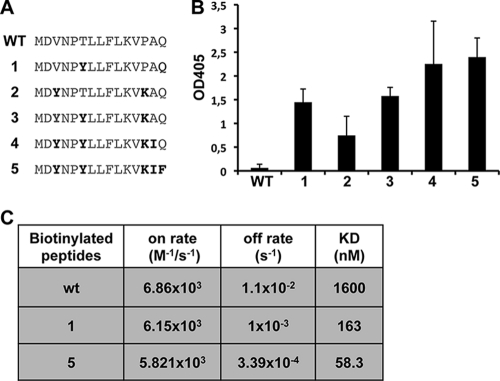
We speculated that simultaneous substitution of several amino acid positions identified by our previous screen might further increase peptide affinity. Since we reasoned that the T6Y substitution within the 310-helix might be most critical in enhancing the affinity of the peptide, we tested additional mutations that might be compatible with this substitution. To this end, we screened another peptide library with PB11-15T6Y as the parent sequence. As expected, the mean signal of the parent sequence was higher than in the PB11-15 wt sequence, increasing from a relative mean value of ca. 26,500 to a relative mean value of ca. 42,700 ± 2,596 (Fig. 2). The reproducibility of this binding assay was further confirmed by equivalent intensities of PB11-15 wt in both peptide library screens (compare the signal intensities of PB11-15 in Fig. 1 and 2). This binding assay also confirmed the necessity of the P5YLLFLK11 sequence, since overall only few substitutions within the core PA-binding region were tolerated. Because of the generally increased affinity of the PB11-15 T6Y peptide, the array allowed much more flexibility in respect to amino acid substitutions in positions 1 to 3 and positions 12 to 15, meaning that only a few substitutions actually resulted in a significant decrease in signal. Importantly, the combination of the affinity-enhancing mutations identified in the first screening assay (V3Y, P13K, A14I, and Q15F) with T6Y is possible without losing binding affinity to PA (Fig. 2). However, these additional substitutions lead to only minor increases in PA binding on the array. This could be due either to a comparatively small linear range of this binding assay or to the fact that the combination of affinity enhancing mutations does not result in a further increase in PA binding. To differentiate between these two possibilities, we determined the affinity of PB11-15 derivatives, in which the V3Y, T6Y, P13K, A14I, and Q15F substitutions were sequentially combined using the ELISA-based binding assay (Fig. 3 A and B). Compared to PB11-15, a single substitution with Y at amino acid position 6 or a double substitution (V3Y and P13K) resulted in a 24- or a 12-fold increase, respectively. Furthermore, combination of all five amino acid substitutions resulted in a 26-fold increase in PA binding compared to PB11-15. These results indicate that the affinity-enhancing mutations identified in the peptide library screening assay do significantly improve binding to PA, although not in a linear manner.
FIG. 2.
Binding characteristics of PA-HA to PB11-15T6Y derivatives in an analogue peptide array. (A) A peptide array with single point mutations in PB11-15T6Y as shown in panel B was incubated with partially purified PA-HA as described in Fig. 1. (B) Quantification of the signal intensities of the peptide array shown in panel A. The PB11-15T6Y sequence is shown horizontally; the individual amino acid substitutions are shown vertically. Labeling is identical with Fig. 1C except for the categories of quantifications: signals of the array are categorized as <35,000 (blue shading), 35,000 to 46,000 (orange shading, includes parent sequences PB11-15T6Y), and >46,000 (red shading representing an increase in signal compared to PB11-15T6Y sequences). The mean of the PB11-15T6Y sequences is 42,675 with a standard deviation of 2,596.
FIG. 3.
PB1 peptides with several affinity enhancing amino acid substitutions show limited increase in PA-binding. (A) Sequences of the biotinylated peptides with the sequential combination of the affinity-enhancing mutations. (B) Binding of PA-HA (A/WSN/33) to immobilized peptides was determined by ELISA. Shown are mean optical density at 405 nm (OD405) values from four independent experiments. The OD405 values represent the increase in bound PA-HA compared to a scrambled peptide. Error bars represent the standard deviations. Note that the PB11-15 wt shows only a slight increase compared to binding background. As a positive control, PB11-25 wt was used (not shown). These settings were chosen because they allow discrimination of high binding efficiencies. (C) Binding kinetics between PA-His and the indicated biotinylated peptides were determined by SPR. KD, dissociation constant.
To determine the equilibrium constants (KD) of selected PB11-15 peptide derivatives, surface plasmon resonance (SPR) analyses were performed using a Biacore system. The biotinylated peptides were immobilized on a streptavidin (SA) sensorchip. Various concentrations of yeast purified PA-His analyte were injected into peptide-immobilized flow cells to obtain SPR sensograms, and the KD values were determined (Fig. 3C). This real-time analysis of the binding characteristics revealed that the affinity-enhancing mutations predominantly decrease the off-rate of the binding, whereas the on-rate hardly changes. This results in 9.8- and 27-fold decreases in the KD from the PB11-15 peptide to the PB11-15T6Y peptide and to the PB11-15V3Y, -T6Y, -P13K, -A14I, and -Q15F peptides, respectively. Thus, the combination of affinity-enhancing amino acid substitutions in PB11-15 significantly improves the binding affinity to PA.
DISCUSSION
Based on a detailed structure-affinity relationship analysis of the PB1-PA binding site of influenza A viruses, we present evidence that the core PA-binding domain of PB1 is essential but not sufficient for binding to PA. We believe that the lack of detectable affinity of PB15-11 to PA is due to the small size of the peptide and the relatively large binding cavity in PA. Therefore, amino acids at the N- and C-terminal ends of the core-binding domain are required to increase the affinity above a certain threshold until stable binding of the core PA-binding domain is achieved.
Several amino acid changes are tolerated outside the core PA-binding domain, most likely since the binding of PA by aa 1, 13, and 14 is mediated by the peptide backbone instead of the side chains (13). However, positions 2 to 4 and positions 11 and 12 can be exchanged only by specific residues, which is partially consistent with specific recognition of PA via their side chains. Positions that do not mediate side chain contacts to PA are probably crucial for preserving the structure of the peptide. Thus, the core PA-binding domain should be extended to aa 2 to 12. We hypothesize that PA binding is initiated by both amino acids outside and within the core PA-binding domain in order to correctly place the core-binding domain into the binding cavity of PA. As a consequence, peptidomimetics solely based on the core-binding domain (PB15-11) are most likely difficult to achieve.
Since the crystal structures suggested unexploited space at amino acid positions 5, 6, and 9 (13), we would have expected an increase in binding upon substitution of these positions by bulky amino acids. However, as mentioned above, only substitution of aa 6 revealed the expected affinity-enhancing effect. Furthermore, since proline at position 5 is probably necessary to preserve the helix structure, we assume that amino acid exchanges at this position are not tolerated. F9 already presents a rather bulky amino acid, and therefore amino acid substitution at this position may even result in decreased affinity.
The majority of affinity enhancing mutations was found outside the core PA-binding domain. Since almost all amino acid substitutions within the core binding domain, with the notable exception of T6Y, abrogated binding to PA, we assume that structural constraints even restrict exchange by very similar amino acids. This is consistent with a very high conservation of amino acid residues within the PA-binding site among all influenza A viruses (5, 22). However, some amino acid positions (1, 2, and 12 to 14) outside the PA-binding domain can be substituted without losing affinity to PA. Interestingly, some of these amino acid changes resulted in a significant increase in PA binding. We speculate that the affinity-enhancing substitutions can directly influence the binding affinity to the PA protein, although an indirect effect by increasing the stability of the 310-helix cannot be excluded. The identified affinity-enhancing mutations predominantly represent hydrophobic amino acids. We therefore assume that hydrophobic interactions contribute the major part of the binding energy. This is consistent with the identification of polar and nonpolar interactions in the crystal structures (8, 13) and the results of a computational approach (11).
Although we could show that the peptide affinity can be significantly improved by substitution with natural amino acids, it is possible that substitution with non-natural amino acids can further enhance the affinity significantly, as shown in the case of an HIV-1 entry inhibitor (3), and simultaneously stabilize the peptide against proteolytic digestion. However, structural stabilization of the 310-helix by fixation in highly helical structures, as recently shown for several bacterial and viral short alpha helices, could increase binding by stabilizing the peptide in its active conformation (7, 20). This could then also lead to a further reduction in size of the peptide.
Antivirally active peptidomimetics based on the PA-binding domain have several advantages. (i) The PA-binding domain is highly conserved among all influenza A viruses, and therefore such peptidomimetics most likely possess antiviral activity against several influenza A virus strains. (ii) Mutation in the PA-binding site leads to significant attenuation of the mutant viruses (21; B. Mänz et al., unpublished), suggesting that escape mutants might not easily emerge under treatment with antivirally active peptidomimetics. (iii) Finally, the PA-PB1 binding site of two influenza A viruses has been resolved by crystal structures (8, 13) and is thus amenable for modeling studies to improve first lead molecules.
Although we do not anticipate the clinical use of the described peptides, we believe that, with the availability of peptides with significantly increased PA-binding affinities, the development of antivirally active peptidomimetics or drug-like molecules seems now feasible.
Acknowledgments
We thank Geoffrey Chase and Johannes Kirchmair for critically reading the manuscript.
K.W. was funded by the Excellence Initiative of the German Research Foundation (GSC-4, Spemann Graduate School). This study was supported by the Deutsche Forschungsgemeinschaft, the European Commission′s Seventh Framework, and FluResearchNet.
Footnotes
Published ahead of print on 6 December 2010.
REFERENCES
- 1.Biswas, S. K., and D. P. Nayak. 1996. Influenza virus polymerase basic protein 1 interacts with influenza virus polymerase basic protein 2 at multiple sites. J. Virol. 70:6716-6722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dias, A., et al. 2009. The cap-snatching endonuclease of influenza virus polymerase resides in the PA subunit. Nature 458:914-918. [DOI] [PubMed] [Google Scholar]
- 3.Eckert, D. M., V. N. Malashkevich, L. H. Hong, P. A. Carr, and P. S. Kim. 1999. Inhibiting HIV-1 entry: discovery of D-peptide inhibitors that target the gp41 coiled-coil pocket. Cell 99:103-115. [DOI] [PubMed] [Google Scholar]
- 4.Frank, R., and H. Overwin. 1996. SPOT synthesis. Epitope analysis with arrays of synthetic peptides prepared on cellulose membranes. Methods Mol. Biol. 66:149-169. [DOI] [PubMed] [Google Scholar]
- 5.Ghanem, A., et al. 2007. Peptide-mediated interference with influenza A virus polymerase. J. Virol. 81:7801-7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guilligay, D., et al. 2008. The structural basis for cap binding by influenza virus polymerase subunit PB2. Nat. Struct. Mol. Biol. 15:500-506. [DOI] [PubMed] [Google Scholar]
- 7.Harrison, R. S., et al. Downsizing human, bacterial, and viral proteins to short water-stable alpha helices that maintain biological potency. Proc. Natl. Acad. Sci. U. S. A. 107:11686-11691. [DOI] [PMC free article] [PubMed]
- 8.He, X., et al. 2008. Crystal structure of the polymerase PA(C)-PB1(N) complex from an avian influenza H5N1 virus. Nature 454:1123-1126. [DOI] [PubMed] [Google Scholar]
- 9.Hemerka, J. N., et al. 2009. Detection and characterization of influenza A virus PA-PB2 interaction through a bimolecular fluorescence complementation assay. J. Virol. 83:3944-3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kosugi, S., et al. 2008. Design of peptide inhibitors for the importin alpha/beta nuclear import pathway by activity-based profiling. Chem. Biol. 15:940-949. [DOI] [PubMed] [Google Scholar]
- 11.Liu, H., and X. Yao. Molecular basis of the interaction for an essential subunit PA-PB1 in influenza virus RNA polymerase: insights from molecular dynamics simulation and free energy calculation. Mol. Pharm. 7:75-85. [DOI] [PubMed]
- 12.Munch, J., et al. 2007. Discovery and optimization of a natural HIV-1 entry inhibitor targeting the gp41 fusion peptide. Cell 129:263-275. [DOI] [PubMed] [Google Scholar]
- 13.Obayashi, E., et al. 2008. The structural basis for an essential subunit interaction in influenza virus RNA polymerase. Nature 454:1127-1131. [DOI] [PubMed] [Google Scholar]
- 14.Ohtsu, Y., Y. Honda, Y. Sakata, H. Kato, and T. Toyoda. 2002. Fine mapping of the subunit binding sites of influenza virus RNA polymerase. Microbiol. Immunol. 46:167-175. [DOI] [PubMed] [Google Scholar]
- 15.Perez, D. R., and R. O. Donis. 1995. A 48-amino-acid region of influenza A virus PB1 protein is sufficient for complex formation with PA. J. Virol. 69:6932-6939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Perez, D. R., and R. O. Donis. 2001. Functional analysis of PA binding by influenza A virus PB1: effects on polymerase activity and viral infectivity. J. Virol. 75:8127-8136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruigrok, R. W., T. Crepin, D. J. Hart, and S. Cusack. Towards an atomic resolution understanding of the influenza virus replication machinery. Curr. Opin. Struct. Biol. 20:104-113. [DOI] [PubMed]
- 18.Salomon, R., and R. G. Webster. 2009. The influenza virus enigma. Cell 136:402-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sugiyama, K., et al. 2009. Structural insight into the essential PB1-PB2 subunit contact of the influenza virus RNA polymerase. EMBO J. 28:1803-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Timmerman, P., J. Beld, W. C. Puijk, and R. H. Meloen. 2005. Rapid and quantitative cyclization of multiple peptide loops onto synthetic scaffolds for structural mimicry of protein surfaces. Chembiochem 6:821-824. [DOI] [PubMed] [Google Scholar]
- 21.Wunderlich, K., et al. Limited compatibility of polymerase subunit interactions in influenza A and B viruses. J. Biol. Chem. 285:16704-16712. [DOI] [PMC free article] [PubMed]
- 22.Wunderlich, K., et al. 2009. Identification of a PA-binding peptide with inhibitory activity against influenza A and B virus replication. PLoS One 4:e7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yuan, P., et al. 2009. Crystal structure of an avian influenza polymerase PA(N) reveals an endonuclease active site. Nature 458:909-913. [DOI] [PubMed] [Google Scholar]